Protein Engineering for Therapeutic Antibodies: From AI-Driven Design to Clinical Applications
This article provides a comprehensive overview of the current landscape and future directions of protein engineering for therapeutic antibodies, tailored for researchers, scientists, and drug development professionals.
Protein Engineering for Therapeutic Antibodies: From AI-Driven Design to Clinical Applications
Abstract
This article provides a comprehensive overview of the current landscape and future directions of protein engineering for therapeutic antibodies, tailored for researchers, scientists, and drug development professionals. It explores the foundational shift from traditional methods to computational and AI-driven design, detailing core methodologies like rational design, directed evolution, and advanced display technologies. The scope extends to troubleshooting critical developability challenges—such as immunogenicity, stability, and aggregation—and concludes with a comparative analysis of clinical and commercial validation, synthesizing key takeaways to guide future R&D strategies in biomedicine.
The New Frontier: How Computational Tools are Reshaping Antibody Therapeutics
The global biologics market has demonstrated exceptional growth, driven significantly by the dominance of monoclonal antibodies (mAbs). The market data underscores a robust expansion, solidifying the commercial and therapeutic impact of antibody-based therapies.
Table 1: Global Biologics Market Size and Projections
| Metric | 2024/2025 Value | 2030/2034 Value | CAGR (Compound Annual Growth Rate) | Source Segment |
|---|---|---|---|---|
| Overall Biologics Market | USD 444.40 billion (2024) [1] | USD 1,144.20 billion (2034) [1] | 9.96% (2025-2034) [1] | - |
| Biologics Manufacturing Market | USD 33.52 billion (2024) [2] | USD 162.51 billion (2034) [2] | 17.1% (2025-2034) [2] | - |
| Monoclonal Antibodies (Product Segment) | USD 274.4 billion (2024) [3] | - | - | - |
| Mammalian Source Systems | 71.34% market share (2024) [3] | - | - | Chinese Hamster Ovary (CHO) cells are the dominant mammalian platform [4] [3]. |
| Microbial Source Systems | 58.78% market share (2024) [1] | - | - | Includes E. coli and yeast [5] [1]. |
The monoclonal antibodies segment is the undisputed leader within the biologics market, accounting for an estimated 56.48% of the market by product in 2024 and generating USD 274.4 billion in revenue [1] [3]. This dominance is attributed to their high specificity, success in clinical applications, and their ability to target diseased cells without harming healthy ones [2] [1]. The oncology therapeutic area commands the largest share at 36.54%, growing at a remarkable CAGR of 13.78%, fueled by the adoption of immunotherapies, antibody-drug conjugates (ADCs), and CAR-T therapies [3].
From a manufacturing perspective, mammalian expression systems, particularly CHO cells, are preferred for producing complex, glycosylated therapeutic antibodies, holding over 71% of the market share by source [2] [3]. This preference stems from their ability to perform human-like post-translational modifications, which are critical for the efficacy and stability of therapeutic proteins [4].
Protein Engineering Strategies for Therapeutic Antibodies
Protein engineering is central to advancing therapeutic antibodies, enabling the enhancement of affinity, stability, and pharmacological properties. These strategies can be broadly classified into directed evolution and rational design.
Directed Evolution and Display Technologies
Directed evolution mimics natural selection in a laboratory setting to isolate antibody variants with desired traits from vast diverse libraries [6].
Phage Display: This pioneering technology, for which the 2018 Nobel Prize in Chemistry was awarded, involves fusing antibody fragments (e.g., scFv, Fab) to a coat protein of a bacteriophage [6]. The process of biopanning allows for the isolation of high-affinity binders through iterative cycles of:
- Incubation: The phage library is exposed to the immobilized target antigen.
- Washing: Non-specific or weak binders are removed.
- Elution: Specifically bound phages are recovered and amplified in E. coli for the next round [6]. Successful drugs like adalimumab (Humira) for rheumatoid arthritis and belimumab (Benlysta) for lupus were discovered and optimized using phage display [6].
Other Display Platforms: Yeast surface display and ribosome/mRNA display are other powerful technologies. Ribosome/mRNA display is a cell-free system that can generate exceptionally large libraries (10^12-10^14 clones) and is not limited by bacterial transformation efficiency, allowing for the isolation of antibodies with picomolar affinities [6].
Rational Design and Engineering Approaches
Rational design employs structural knowledge and computational tools to make precise modifications to antibody sequences [7].
Affinity and Stability Optimization: Site-directed mutagenesis is used to improve antibody characteristics. For instance, computational tools like Spatial Aggregation Propensity (SAP) can identify regions prone to aggregation, guiding mutations that enhance stability [7]. Substituting free cysteine residues with serine prevents unwanted disulfide bond formation and oxidation, a strategy used in approved drugs like pegfilgrastim (Neulasta) [7].
Fc Engineering for Enhanced Pharmacokinetics: The circulating half-life of antibodies is modulated by their interaction with the neonatal Fc receptor (FcRn). Introducing specific point mutations (e.g., M428L/N434S, known as the "LS" variant) in the Fc region increases antibody half-life by enhancing FcRn binding at acidic pH, promoting recycling over lysosomal degradation. This engineering approach is utilized in ravulizumab (Ultomiris), which has an extended dosing interval compared to its predecessor [7].
Reducing Immunogenicity: A key goal in protein engineering is to minimize immune responses. Techniques like humanization—replacing murine sequences with human counterparts in antibodies derived from mice—drastically reduce immunogenicity [6]. Further deimmunization strategies involve identifying and mutating potential T-cell epitopes within the antibody sequence [7].
Formulating Bispecifics and Antibody-Drug Conjugates (ADCs): Engineering has enabled the creation of bispecific antibodies that can engage two different targets simultaneously, and ADCs that deliver potent cytotoxic drugs directly to cancer cells, maximizing efficacy and minimizing systemic toxicity [3].
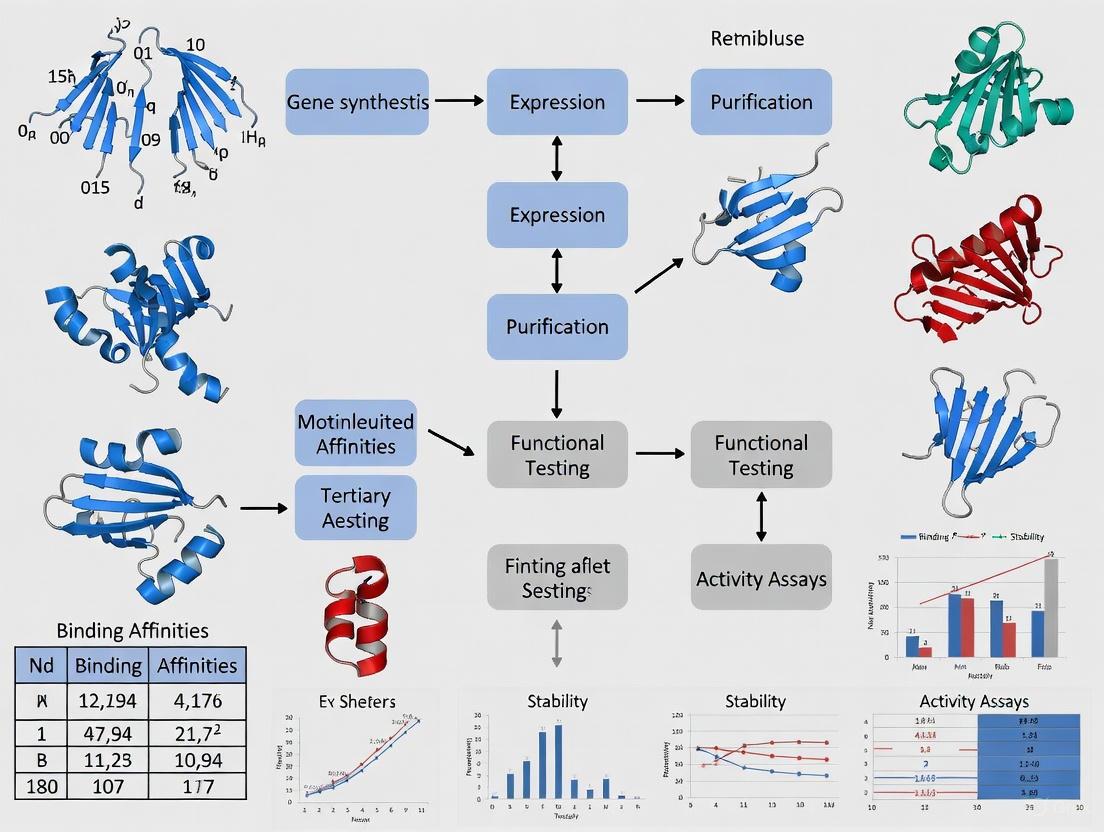
The following diagram illustrates the logical workflow and key decision points in the antibody engineering and development process.
Application Notes: Detailed Experimental Protocols
This section provides detailed methodologies for key experiments in antibody development, from discovery to biophysical characterization.
Protocol 1: Phage Display Biopanning for Antibody Selection
Objective: To isolate antigen-specific antibody fragments (scFv or Fab) from a naive or immune phage display library through iterative selection rounds [6].
Materials:
- Phage display library (e.g., human scFv library)
- Target antigen (recombinant protein, purified)
- Immunotubes or ELISA plates for immobilization
- Washing buffers (PBS with 0.1% Tween-20, PBS alone)
- Elution buffer (0.1 M Glycine-HCl, pH 2.2, or Triethylamine)
- Neutralization buffer (1 M Tris-HCl, pH 9.1)
- E. coli strains for infection (e.g., TG1)
- Culture media (2xYT with appropriate antibiotics)
Procedure:
- Antigen Coating: Coat an immunotube or well with 1-10 µg/mL of target antigen in PBS overnight at 4°C. Include a negative control well without antigen.
- Blocking: Block the coated surface with 2-4% Marvel/PBS (or BSA/PBS) for 1-2 hours at room temperature to prevent non-specific binding.
- Phage Incubation: Incubate the phage library (10^12-10^13 phage particles) in blocking buffer for 1-2 hours to allow binding.
- Washing: Remove unbound phages by rigorous washing.
- First Round: 10-20 washes with PBS/0.1% Tween-20, followed by 10-20 washes with PBS.
- Subsequent Rounds: Increase stringency by increasing the number of washes or Tween concentration.
- Elution: Recover specifically bound phages using two methods:
- Acidic Elution: Add 0.5-1 mL of 0.1 M Glycine-HCl (pH 2.2) for 5-15 minutes. Immediately neutralize with 0.5-1 mL of 1 M Tris-HCl (pH 9.1).
- Competitive Elution (Alternative): Incubate with excess soluble antigen for 30-60 minutes.
- Amplification: Infect log-phase E. coli TG1 cells with the eluted phages. Culture the infected cells to amplify the phage pool for the next selection round. Helper phages are added to rescue the phagemid particles.
- Titration: Determine the input and output phage titers after each round to monitor enrichment. A significant increase in output titer on antigen-coated wells compared to control indicates successful selection.
- Repeat: Typically, 3-4 rounds of biopanning are performed to sufficiently enrich for high-affinity binders.
- Screening: After the final round, isolate single clones for screening (e.g., via ELISA) to identify antigen-positive antibody fragments.
Protocol 2: Site-Directed Mutagenesis for Fc Engineering
Objective: To introduce specific point mutations (e.g., M428L/N434S) into the Fc region of an IgG antibody to enhance FcRn binding and extend serum half-life [7].
Materials:
- Plasmid DNA containing the gene for the IgG antibody of interest.
- High-fidelity DNA polymerase (e.g., PfuUltra).
- DpnI restriction enzyme.
- Mutagenic primers designed for the desired mutation.
- Competent E. coli cells.
Procedure:
- Primer Design: Design and synthesize complementary forward and reverse primers (25-45 bases) that contain the desired mutation in the center, flanked by 10-15 correct nucleotides on each side.
- PCR Amplification: Set up a PCR reaction using the plasmid as a template and the mutagenic primers. The high-fidelity polymerase will amplify the entire plasmid, incorporating the mutation.
- Typical Thermocycler Conditions:
- Initial Denaturation: 95°C for 2 minutes.
- 18 Cycles:
- Denature: 95°C for 30 seconds.
- Anneal: 55-65°C for 1 minute.
- Extend: 72°C for 1-2 minutes per kb of plasmid length.
- Final Extension: 72°C for 10 minutes.
- Typical Thermocycler Conditions:
- DpnI Digestion: After PCR, treat the reaction mixture with DpnI for 1 hour at 37°C. DpnI cleaves the methylated parental DNA template, leaving the newly synthesized, mutated DNA strand intact.
- Transformation: Transform the DpnI-treated DNA into competent E. coli cells.
- Screening and Sequencing: Plate the cells and pick colonies. Isolate plasmid DNA and verify the introduction of the correct mutation by DNA sequencing.
- Protein Expression: Transfect the validated plasmid into mammalian expression cells (e.g., HEK293 or CHO) for transient or stable expression of the engineered full-length antibody.
Protocol 3: Tangential Flow Filtration (TFF) for mAb Purification
Objective: To concentrate and buffer-exchange a clarified cell culture harvest containing a monoclonal antibody, as a key step in downstream processing [4].
Materials:
- Clarified cell culture supernatant.
- Tangential Flow Filtration system with a peristaltic pump.
- Pellicon cassette or hollow fiber module (appropriate MWCO, e.g., 30 kDa for mAbs).
- Diafiltration buffer (e.g., PBS for final formulation, or a different buffer for intermediate purification).
- Conductivity and pH meters.
Procedure:
- System Setup and Equilibration: Assemble the TFF system according to the manufacturer's instructions. Flush the system and membrane with water, then equilibrate with diafiltration buffer.
- Concentration:
- Load the clarified harvest into the feed reservoir.
- Recirculate the fluid, applying pressure to force buffer and small molecules (permeate) through the membrane while retaining the antibody (retentate).
- Continue until the desired concentration factor (e.g., 10x) is achieved.
- Diafiltration (Buffer Exchange):
- Once concentrated, begin adding diafiltration buffer to the retentate reservoir at the same rate as the permeate is removed. This process washes out small impurities and exchanges the buffer.
- Typically, 5-10 volume exchanges are performed to ensure >99% buffer exchange.
- Final Recovery:
- After diafiltration, recover the concentrated and buffer-exchanged retentate.
- Flush the system with a small volume of buffer to maximize product recovery.
- Cleaning and Storage: Clean the TFF membrane immediately after use according to the manufacturer's protocol (e.g., with NaOH solution) and store in an appropriate preservative.
The following workflow diagram maps the key stages of the antibody biomanufacturing process.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 2: Key Reagents and Materials for Antibody Research and Development
| Item | Function/Application | Key Considerations |
|---|---|---|
| CHO (Chinese Hamster Ovary) Cells | Mammalian host cell line for producing full-length, glycosylated therapeutic antibodies [5] [4]. | Capable of human-like post-translational modifications; requires complex media and controlled bioreactor conditions [3]. |
| Phagemid Vectors | Plasmids used in phage display to display antibody fragments on the surface of filamentous phage (e.g., M13) [6]. | Contains both bacterial and phage origins of replication, antibiotic resistance marker, and a fusion gene for a coat protein (pIII or pVIII) [6]. |
| Protein A/G/L Chromatography Resins | Affinity chromatography matrices for purifying antibodies and Fc-fusion proteins from complex mixtures like cell culture supernatant [4]. | Binds with high specificity to the Fc region of antibodies; a cornerstone of downstream purification processes [4]. |
| Single-Use Bioreactors | Disposable bags used for cell culture in upstream biomanufacturing [4] [3]. | Eliminates cross-contamination risk and reduces cleaning validation; enables flexible and scalable production [4] [3]. |
| TFB (Transformation Buffer) | Chemically competent E. coli cells used for high-efficiency transformation following mutagenesis or library construction [6]. | Essential for cloning steps, library amplification, and plasmid propagation. |
| Ethylenethiourea-d4 | Ethylenethiourea-d4, CAS:352431-28-8, MF:C3H6N2S, MW:106.19 g/mol | Chemical Reagent |
| 2,3-Dichlorobenzoic acid-13C | 2,3-Dichlorobenzoic acid-13C, CAS:1184971-82-1, MF:C7H4Cl2O2, MW:192.00 g/mol | Chemical Reagent |
The field of therapeutic antibody development has undergone a profound transformation, evolving from biologically-driven methods reliant on the immune systems of animals to sophisticated computational design performed entirely in silico [8]. This paradigm shift is rooted in protein engineering, which applies principles of rational design, directed evolution, and, most recently, artificial intelligence (AI) to create antibodies with enhanced specificity, efficacy, and safety profiles [9]. The journey began with hybridoma technology, which enabled the production of murine monoclonal antibodies, and progressed through chimeric, humanized, and fully human antibodies to mitigate immunogenicity [8]. Today, the integration of AI and machine learning (ML) is reshaping the discovery landscape, dramatically accelerating timelines and enabling the targeting of previously "undruggable" epitopes [10] [11]. This article details the key methodologies driving this evolution, providing application notes and experimental protocols for researchers and drug development professionals.
Evolution of Antibody Discovery Platforms
The following table summarizes the quantitative progression of key technologies that have defined the antibody discovery landscape, highlighting the transition from biological to computational dominance.
Table 1: Key Metrics in the Evolution of Antibody Discovery Platforms
| Discovery Platform | Timeline (First Approved Drug) | Key Advantage | Key Limitation | Impact on Discovery Timelines |
|---|---|---|---|---|
| Hybridoma Technology [8] | 1980s (Muromonab-CD3, 1986) | High-affinity, native paired antibodies | Murine origin leads to immunogenicity (HAMA response) | 12-18 months |
| Phage Display [8] [12] | 1990s (Adalimumab, 2002) | Bypasses immunization; fully human antibodies | Limited to smaller constructs (e.g., scFv); bacterial folding machinery | 9-12 months |
| Transgenic Mice [8] | 2000s (Panitumumab, 2006) | Fully human antibodies with native pairing | Complex and costly platform development | 10-14 months |
| Single B-Cell Screening [8] | 2010s | Preserves native heavy-light chain pairing; rapid for infectious diseases | Requires specific patient/donor samples | 6-9 months |
| AI/ML & In Silico Design [13] [11] | 2020s (Clinical candidates, e.g., Imneskibart) | De novo design; targets "undruggable" proteins; minimal immunogenicity | Requires high-quality, large-scale data for training | < 6 weeks for initial candidate generation |
The market dynamics reflect this technological evolution. The global antibody discovery market, valued at USD 2.06 billion in 2025, is projected to grow at a CAGR of 9.8% to reach USD 5.25 billion by 2035 [12]. A significant trend is the segment growth by antibody type, where fully human antibodies are poised to dominate due to demand for reduced immunogenicity and enhanced efficacy in personalized medicine [12].
Table 2: Antibody Discovery Market Growth by Type (2025-2035 Projection)
| Antibody Type | Key Driver | Projected Market Dominance |
|---|---|---|
| Murine Antibody | Historical use; regulatory familiarity | Ceding ground |
| Chimeric Antibody | Reduced immunogenicity vs. murine | Stable niche |
| Humanized Antibody | Further reduced immunogenicity | Widely used |
| Human Antibody | Minimal immunogenicity; superior efficacy | Projected market leader by 2035 |
Application Notes & Experimental Protocols
Protocol 1: Traditional Murine Hybridoma Generation for Monoclonal Antibody Production
This protocol outlines the classic method for generating monoclonal antibodies, which remains a foundational technique for obtaining antibodies with native pairing [8].
Reagents & Equipment:
- Immunogen (purified protein, peptide, or cells)
- Adjuvant (e.g., Complete and Incomplete Freund's Adjuvant)
- Female BALB/c mice (6-8 weeks old)
- Myeloma cell line (e.g., SP2/0 or P3X63Ag8.653)
- Polyethylene glycol (PEG) solution for fusion
- HAT (Hypoxanthine-Aminopterin-Thymidine) selection medium
- HT (Hypoxanthine-Thymidine) medium
- ELISA plates and coating buffers for screening
- Cell culture flasks and COâ‚‚ incubator
Procedure:
- Immunization: Emulsify 10-100 µg of immunogen with an equal volume of Freund's Complete Adjuvant. Administer the emulsion intraperitoneally to mice. Perform two booster immunizations at 2-3 week intervals using immunogen emulsified with Incomplete Freund's Adjuvant. Test serum titers by ELISA 7-10 days after the final boost.
- Cell Preparation & Fusion: 3-4 days after a final booster immunization, sacrifice the mouse with the highest serum titer and aseptically remove the spleen. Prepare a single-cell suspension of splenocytes in serum-free medium. Culture the myeloma cells to ensure they are in log-phase growth. Mix splenocytes and myeloma cells at a 10:1 ratio and pellet by centrifugation. Slowly add 1 mL of 50% PEG 1500 to the cell pellet over 1 minute with gentle agitation. Slowly dilute the PEG with serum-free medium over 5-7 minutes.
- HAT Selection & Cloning: Resuspend the fused cells in HAT medium supplemented with 20% Fetal Bovine Serum (FBS) and plate into 96-well tissue culture plates. Incubate at 37°C in a 5% CO₂ humidified incubator. Feed cells with HAT medium every 3-4 days. After 10-14 days, screen supernatant from wells with hybridoma growth for desired antigen specificity by ELISA. Perform at least two rounds of limiting dilution subcloning of positive wells to ensure monoclonality, using HT medium during the first subcloning.
- Expansion & Characterization: Expand stable, monoclonal hybridoma lines. Isotype the produced antibody. Cryopreserve positive clones in liquid nitrogen for long-term storage.
Protocol 2: AI-DrivenDe NovoAntibody Design andIn VitroValidation
This protocol describes a modern computational workflow for generating and validating novel antibody sequences, leveraging powerful AI models like RFdiffusion and ProteinMPNN [10] [11].
Reagents & Equipment:
- High-performance computing (HPC) cluster with GPU acceleration
- AI/ML Software: RFdiffusion [10], ProteinMPNN [10], AlphaFold2 [10] or similar (ESM-IF, Chai-2 [11])
- Target antigen structure (experimental or AlphaFold2-predicted)
- Gene synthesis service
- Mammalian expression system (e.g., HEK293 or CHO cells)
- Biacore or Octet system for binding kinetics
- UPLC-SEC for aggregation analysis
Procedure:
- Target Featurization and Scaffold Specification: Input the 3D structure of the target antigen (e.g., from PDB or an AlphaFold2 prediction) into the design platform. Define design constraints, including the desired epitope region and any specific structural scaffolds for the antibody (e.g., a particular IgG subclass framework).
- Paratope and Backbone Generation: Use a diffusion model (e.g., RFdiffusion) to generate novel protein backbones that are complementary to the target epitope. This model samples the conformational landscape to create binders inspired by, but distinct from, natural antibodies [10].
- Sequence Design with Inverse Folding: For each generated backbone, use an inverse folding algorithm such as ProteinMPNN or ESM-IF to design a amino acid sequence that is most likely to fold into that specific structure. These tools achieve a high sequence recovery rate (~53%), significantly improving on older physics-based methods [10].
- In Silico Ranking and Filtering: Score the designed antibody sequences using models that predict affinity, stability, and developability (e.g., solubility, low polyspecificity). Filter the thousands of in silico designs down to the top 10-50 candidates for synthesis based on these scores. This step can achieve a hit rate of 15.5% or higher in subsequent testing [11].
- Gene Synthesis, Expression, and In Vitro Validation: Outsource the coding sequences of the top candidates for gene synthesis. Clone the genes into an mammalian expression vector, transiently transfect HEK293 cells, and purify the expressed antibodies using protein A/G chromatography. Validate the designs experimentally:
- Binding Kinetics: Determine affinity (KD) and kinetics (kon, koff) using surface plasmon resonance (SPR) or bio-layer interferometry (BLI).
- Specificity: Test binding to the target antigen versus unrelated proteins via ELISA.
- Developability: Assess stability under stressed conditions (e.g., thermal shift assay) and measure aggregation propensity by UPLC-SEC.
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for Antibody Discovery and Engineering
| Research Reagent / Solution | Function & Application | Key Features |
|---|---|---|
| Phage Display Library [8] [12] | A diverse collection of bacteriophages displaying antibody fragments (e.g., scFv, Fab) used for in vitro selection of binders. | High diversity (>10^10 unique clones); enables discovery without animal immunization. |
| Hybridoma Fusion Partner Cell Line [8] | An immortal myeloma cell line (e.g., SP2/0) deficient in HGPRT, used for fusion with B-cells to create immortal hybridomas. | Enzyme-deficient for selection with HAT medium; high fusion efficiency. |
| HAT/HT Selection Media [8] | A critical cell culture supplement for selecting successfully fused hybridomas and eliminating unfused myeloma cells post-electrofusion. | Contains Hypoxanthine, Aminopterin, and Thymidine; Aminopterin blocks the de novo synthesis pathway. |
| AI-Driven Drug Discovery Platform (e.g., BioNeMo, Chai Discovery) [13] [11] | Integrated computational environments using generative AI and ML models for de novo antibody design and optimization. | Models like RFdiffusion and ProteinMPNN; enables in silico affinity maturation and developability prediction. |
| Microfluidic Single B-Cell Screening System [8] [13] | A high-throughput platform for isolating, analyzing, and sequencing individual antigen-specific B-cells from immunized animals or convalescent patients. | Preserves native VH-VL pairing; allows for rapid recovery of full-length antibody sequences. |
| 2-Carboxyphenol-d4 | Salicylic Acid-d4 | Certified Reference Standard | Salicylic Acid-d4 is an internal standard for LC/MS or GC/MS applications in pharmaceutical and clinical research. This product is for research use only and not for human use. |
| 6-Chloro-1,3,5-triazine-2,4-diamine-13C3 | 6-Chloro-1,3,5-triazine-2,4-diamine-13C3, CAS:1216850-33-7, MF:C3H4ClN5, MW:148.53 g/mol | Chemical Reagent |
Workflow Visualization
The following diagrams, created using Graphviz DOT language, illustrate the logical relationships and experimental workflows central to the evolution of antibody discovery.
Diagram Title: Traditional Hybridoma Generation Workflow
Diagram Title: Modern AI-Driven Antibody Discovery Workflow
Diagram Title: The Shift in Antibody Discovery Paradigms
The discovery and optimization of therapeutic antibodies, a cornerstone of modern biologics, have traditionally relied on experimental methods such as animal immunization and display technologies. These approaches, while effective, are often time-consuming, labor-intensive, and limited in their ability to explore the vast sequence space of antibodies [10]. The field is now undergoing a profound transformation driven by computational advances. The integration of high-accuracy protein structure prediction tools like AlphaFold2 with sophisticated machine learning (ML) models is breaking long-standing design barriers, enabling the rapid in silico design of antibodies with tailored properties [14] [15]. This document details the specific applications and experimental protocols underpinning this computational leap, providing a framework for its implementation in therapeutic antibody research.
Core Computational Technologies
Revolutionizing Structure Prediction with AlphaFold2
AlphaFold2 represents a fundamental shift in computational biology, providing the first method capable of regularly predicting protein structures with atomic accuracy, even in the absence of homologous structures [16]. Its architecture is uniquely suited to addressing challenges in antibody design.
Key Architectural Innovations of AlphaFold2:
- Evoformer: A novel neural network block that processes inputs through repeated layers, operating on both a multiple sequence alignment (MSA) representation and a pair representation. It enables continuous information exchange between the evolutionary (MSA) and spatial (pair) data streams [16].
- Structure Module: This module introduces an explicit 3D structure, initialized from a trivial state and rapidly refined into a highly accurate atomic model. It employs an equivariant transformer to reason about side-chain atoms and uses a loss function that emphasizes the orientational correctness of residues [16].
- Iterative Refinement (Recycling): The network's output is recursively fed back into the same modules, allowing for iterative refinement that significantly enhances prediction accuracy [16].
For antibody researchers, the resulting explosion of reliably predicted structures—from approximately 200,000 in the Protein Data Bank (PDB) to over 200 million in the AlphaFold database—has dramatically expanded the number of viable starting templates for design projects [10].
Machine Learning for Sequence Design and Optimization
Complementing the structural revolution, machine learning models are revolutionizing how antibody sequences are designed and optimized. These approaches can be broadly categorized as follows:
- Unsupervised & Generative Models: Trained on massive datasets of protein or antibody sequences (e.g., from the Observed Antibody Space (OAS) database), these models learn the underlying principles of "natural" or "fit" sequences [14] [17]. They can then generate novel antibody sequences with native-like biophysical properties, such as high stability. For instance, autoregressive models have been used to design nanobody libraries with 1000-fold greater expression than conventional methods [14].
- Supervised & Bayesian Optimization Models: These models are fine-tuned on experimental data (e.g., binding affinity measurements from yeast display) to predict specific extrinsic properties. A notable end-to-end Bayesian framework has been demonstrated to design single-chain variable fragment (scFv) libraries where 99% of the variants showed improved binding over the initial candidate, representing a 28.7-fold improvement over directed evolution in a head-to-head comparison [15].
Table 1: Key Machine Learning Model Types in Antibody Engineering
| Model Type | Training Data | Primary Function | Example Application/Outcome |
|---|---|---|---|
| Protein Language Models (e.g., BERT) | Broad protein sequence databases (e.g., UniRef) | Predict evolutionarily likely mutations | Affinity maturation of anti-viral antibodies, achieving up to 160-fold affinity improvement [14] |
| Antibody-Specific Language Models | Antibody-specific sequence repertoires (e.g., OAS) | Generate stable, native-like antibody sequences | Design of highly stable nanobody libraries with high expression yields [14] |
| Bayesian Optimization Models | High-throughput binding affinity data | Design high-affinity binders with uncertainty quantification | Generation of diverse scFv libraries where nearly all variants are improvements over the parent candidate [15] |
Application Notes: Integrated Computational Workflows
Workflow 1: De Novo Binder Design
This protocol describes a structure-centric approach for designing novel antibody binders from scratch, leveraging structure prediction and de novo design tools.
Diagram 1: De novo binder design workflow
Detailed Protocol:
- Generate Target Antigen Structure: For antigens with unknown structure, use AlphaFold2 to predict a high-confidence 3D model [10].
- De Novo Backbone Design: Use a diffusion-based model like RFDiffusion to generate novel protein backbones that are structurally complementary to the target antigen's epitope. The process can be constrained with a given active site or binding partner motif [10].
- Sequence Design for Backbone: Using the generated backbone as a fixed scaffold, employ inverse folding tools such as ProteinMPNN or ESM-IF to design a sequence that is most likely to fold into that structure. These tools use graph-based architectures to model residue microenvironments and achieve sequence recovery rates of over 50% [10].
- In silico Validation:
- Self-Consistency Check: Predict the structure of the designed sequence using AlphaFold2 and align it to the designed backbone. A high degree of similarity increases confidence.
- Interaction Validation: Use AlphaFold-Multimer or similar tools to co-fold the designed binder with the target antigen. While this remains a challenging task, it can provide insights into the binding mode and interface [10].
- Experimental Validation: Synthesize the top-ranking designs and characterize them experimentally for expression, stability, and binding affinity.
Workflow 2: ML-Driven Affinity Maturation
This protocol outlines a data-driven, sequence-centric approach for enhancing the affinity of an existing antibody candidate, bypassing the need for explicit structural information.
Diagram 2: ML-driven affinity maturation workflow
Detailed Protocol:
- Generate High-Throughput Training Data:
- Method: Use a yeast mating assay or phage display to screen a library of random mutants (e.g., k=1, 2, 3 mutations within CDRs) of your candidate antibody.
- Output: Quantify binding affinity for tens of thousands of variants, creating a labeled dataset linking sequence to function [15].
- Pre-train a Language Model: Utilize a transformer-based language model (e.g., BERT) that has been pre-trained on a large corpus of protein sequences (Pfam) or, preferably, antibody-specific sequences (OAS database). This provides the model with a foundational understanding of protein biochemistry [15].
- Supervised Fine-Tuning: Fine-tune the pre-trained language model on the experimental binding data generated in Step 1. The model learns to predict binding affinity from sequence. Use an ensemble or Gaussian Process method to provide uncertainty estimates for the predictions [15].
- Bayesian Optimization for Library Design:
- Construct a fitness landscape that maps any antibody sequence to its posterior probability of having better affinity than the parent.
- Use sampling algorithms (e.g., Gibbs sampling, Genetic Algorithms) to explore this landscape and propose sequences that maximize the fitness function. Gibbs sampling is particularly effective for generating highly diverse candidates [15].
- Experimental Validation: Synthesize the computationally designed library (e.g., ~10^4 sequences) and test it empirically using the same high-throughput assay from Step 1.
Table 2: Key Performance Metrics from Recent ML-Driven Antibody Engineering Studies
| Study Focus | Method | Key Comparative Result | Diversity Outcome |
|---|---|---|---|
| scFv Affinity Maturation [15] | Bayesian Optimization + Language Models | 28.7-fold improvement in binding over directed evolution | >99% of designed scFvs were improvements over candidate |
| Anti-Virus Antibody Affinity [14] | Protein Language Models (Unsupervised) | Up to 160-fold binding affinity improvement | Successful with small sets of mutations (~10-20) |
| Nanobody Stability & Expression [14] | Generative Autoregressive Model | 1000-fold greater library expression than conventional design | Generated diverse set of ~185,000 CDR3 sequences |
Table 3: Key Reagents and Resources for Computational Antibody Design
| Resource Category | Name | Function & Application |
|---|---|---|
| Key Software & Algorithms | AlphaFold2 / AlphaFold-Multimer [16] [10] | Predicts 3D structures of monomeric proteins and protein complexes from sequence. |
| RFDiffusion [10] | Generative model for creating novel protein backbones, constrained by motifs or binding partners. | |
| ProteinMPNN / ESM-IF [10] | Inverse folding tools that design sequences for a given protein backbone structure. | |
| Rosetta [10] | Suite for biomolecular modeling and design, using energy functions for structure prediction and design. | |
| Critical Databases | Structural Antibody Database (SAbDab) [17] | Curated, up-to-date repository of all publicly available antibody and nanobody structures. |
| Observed Antibody Space (OAS) [15] [17] | Massive collection of annotated antibody sequence data from next-generation sequencing studies. | |
| Theraputic Antibody Databases (e.g., TABS, SAbDab-Therapeutic) [17] | Curate information on clinically investigated and approved antibody therapeutics. | |
| Experimental Systems for Data Generation | Yeast Display [15] | High-throughput platform for screening antibody libraries and generating quantitative binding data for ML models. |
| Phage Display [10] [15] | Well-established technology for selecting binders from large libraries, often used for initial candidate discovery. |
Critical Challenges and Future Directions
Despite significant progress, several challenges must be addressed to fully realize the potential of computational antibody design.
- Data Quality and Volume: A primary bottleneck is the scarcity of large, diverse, and high-quality experimental datasets for training and validation. Current datasets are often small and skewed (e.g., over half the mutations in one major database are alanine scans), leading models to overfit and fail to generalize [18]. Robust AI models require not just more data, but more varied data—with estimates suggesting a need for at least 90,000 experimentally measured mutations for generalizable predictions [18].
- Antibody-Antigen Co-Folding: Accurately predicting the complex structure of an antibody bound to its antigen remains a formidable challenge. Performance of tools like AlphaFold-Multimer on antibody-antigen interactions is still variable and requires improvement [10].
- Multiparameter Optimization: While affinity is a key goal, therapeutic antibodies also require optimal developability properties (e.g., low immunogenicity, high solubility, low viscosity). Integrating predictive models for these multiple parameters into a single design framework is an active area of research.
The convergence of accurate structure prediction, powerful generative models, and high-throughput experimental validation is ushering in a new era for therapeutic antibody development. By adopting the protocols and resources outlined in this document, researchers can leverage these computational leaps to accelerate the design of better, safer, and more effective antibody therapeutics.
The field of protein engineering is undergoing a revolutionary transformation through the integration of generative artificial intelligence (AI). These advanced computational tools enable the de novo design of protein structures and functions with precision that now rivals or even surpasses nature's capabilities [19]. For researchers focused on therapeutic antibody development, two platforms have emerged as particularly transformative: RFdiffusion for protein backbone generation and ProteinMPNN for sequence design. These tools operate synergistically within a comprehensive design pipeline that begins with structural blueprints and concludes with functionally optimized sequences, offering unprecedented control over protein therapeutics design.
RFdiffusion represents a fundamental advancement as a generative model for protein backbones based on denoising diffusion probabilistic models. By fine-tuning the RoseTTAFold structure prediction network on protein structure denoising tasks, RFdiffusion achieves outstanding performance across diverse design challenges including unconditional protein monomer design, protein binder design, and symmetric oligomer design [20]. The methodology initializes random residue frames and iteratively denoises them through multiple steps, progressively refining the structure toward realistic protein architectures. This approach enables researchers to generate elaborate protein structures with minimal overall structural similarity to naturally occurring proteins in the Protein Data Bank, demonstrating considerable generalization beyond known structural space [20].
Complementing RFdiffusion, ProteinMPNN (Protein Message Passing Neural Network) serves as a deep-learning-based protein sequence design method that operates on fixed protein backbones. Whereas traditional physicochemical energy functions often struggle with the combinatorial complexity of sequence space, ProteinMPNN leverages neural network architectures to rapidly generate optimal sequences for given structural scaffolds. The network treats protein residues as nodes in a graph, with edges defined by Cα–Cα distances, and encodes backbone geometry through pairwise distances between key atoms [21]. This architecture enables high-speed sequence design that scales linearly with protein length, making it practical for designing large protein complexes and libraries for experimental screening.
RFdiffusion: Architectural Framework and Mechanisms
RFdiffusion employs a sophisticated diffusion model framework that builds upon the foundational principles of RoseTTAFold. The system represents protein backbones using a frame representation that includes a Cα coordinate and N-Cα-C rigid orientation for each residue [20]. During training, the model learns to reverse a structured noising process applied to protein structures from the Protein Data Bank. The noising schedule corrupts structures over up to 200 steps through two primary mechanisms: Cα coordinates are perturbed with 3D Gaussian noise, while residue orientations are disturbed using Brownian motion on the manifold of rotation matrices [20].
A critical innovation in RFdiffusion is its implementation of self-conditioning, a training strategy inspired by recycling in AlphaFold2. Unlike canonical diffusion models where predictions at each timestep are independent, self-conditioning allows the model to condition on previous predictions between timesteps [20]. This approach significantly enhances performance on in silico benchmarks for both conditional and unconditional protein design tasks by increasing the coherence of predictions within denoising trajectories. The model is trained using a mean-squared error loss between frame predictions and true protein structures without alignment, which promotes continuity of the global coordinate frame between timesteps—a crucial distinction from the frame aligned point error loss used in standard RoseTTAFold training [20].
For therapeutic antibody design, researchers have developed a specialized version of RFdiffusion fine-tuned specifically on antibody complex structures [22]. This variant incorporates several key modifications: (1) the antibody framework sequence and structure can be provided as conditioning input, enabling designs that maintain stable scaffold regions while innovating in complementarity-determining regions (CDRs); (2) the framework structure is provided in a global-frame-invariant manner using the template track of RFdiffusion, which encodes pairwise distances and dihedral angles as a 2D matrix; and (3) one-hot encoded "hotspot" features allow specification of epitope residues, directing the designed CDRs toward targeted binding regions [22].
ProteinMPNN and Its Specialized Variants
ProteinMPNN represents a fundamental shift from physics-based sequence design methods toward deep learning approaches. The core architecture employs an encoder-decoder framework where protein residues are treated as nodes in a graph, with edges connecting residues based on Cα–Cα distances [21]. The encoder processes input features through multiple message-passing layers that capture both local and long-range interactions within the protein structure. A key advantage of ProteinMPNN is its use of random autoregressive decoding, which enables the design of symmetric complexes and multistate proteins by sequentially generating residues while accounting for previously designed positions [21].
The introduction of LigandMPNN has extended ProteinMPNN's capabilities to explicitly model interactions with nonprotein components—a critical requirement for designing functional antibodies, enzymes, and biosensors [21]. LigandMPNN incorporates several architectural innovations: (1) a protein-ligand graph with edges between protein residues and nearby ligand atoms (within ~10Å); (2) fully connected ligand graphs for each protein residue to enrich representation of ligand geometry; and (3) additional encoder layers dedicated to processing protein-ligand interactions [21]. This architecture significantly outperforms both Rosetta and standard ProteinMPNN on native backbone sequence recovery for residues interacting with small molecules (63.3% versus 50.4% and 50.5%), nucleotides (50.5% versus 35.2% and 34.0%), and metals (77.5% versus 36.0% and 40.6%) [21].
For therapeutic applications, CAPE-Beam represents another specialized decoding strategy for ProteinMPNN that addresses immunogenicity concerns [23]. This approach minimizes cytotoxic T-lymphocyte immunogenicity risk by constraining designs to only include peptide fragments (kmers) that are either predicted not to be presented to CTLs or are subject to central tolerance mechanisms. This capability is particularly valuable for engineering therapeutic proteins intended for chronic administration or broad patient populations where immune responses could limit efficacy or cause adverse effects [23].
Figure 1: Integrated computational workflow for de novo antibody design using RFdiffusion and ProteinMPNN. The pipeline begins with structural generation, proceeds through sequence optimization, and concludes with validation and affinity maturation.
Quantitative Performance Benchmarks
Structure Generation and Sequence Recovery Metrics
Table 1: Performance comparison of RFdiffusion and ProteinMPNN against established methods across key protein design tasks
| Design Task | Method | Performance Metric | Result | Reference |
|---|---|---|---|---|
| Small molecule-binding proteins | LigandMPNN | Sequence recovery (residues within 5Ã…) | 63.3% | [21] |
| Small molecule-binding proteins | ProteinMPNN | Sequence recovery (residues within 5Ã…) | 50.4% | [21] |
| Small molecule-binding proteins | Rosetta (genpot) | Sequence recovery (residues within 5Ã…) | 50.4% | [21] |
| Nucleotide-binding proteins | LigandMPNN | Sequence recovery (residues within 5Ã…) | 50.5% | [21] |
| Nucleotide-binding proteins | ProteinMPNN | Sequence recovery (residues within 5Ã…) | 34.0% | [21] |
| Nucleotide-binding proteins | Rosetta (DNA-optimized) | Sequence recovery (residues within 5Ã…) | 35.2% | [21] |
| Metal-binding proteins | LigandMPNN | Sequence recovery (residues within 5Ã…) | 77.5% | [21] |
| Metal-binding proteins | ProteinMPNN | Sequence recovery (residues within 5Ã…) | 40.6% | [21] |
| Metal-binding proteins | Rosetta | Sequence recovery (residues within 5Ã…) | 36.0% | [21] |
| Unconditional monomer generation | RFdiffusion | In silico success rate* | High (300-600 residue proteins) | [20] |
| Antibody design | Fine-tuned RFdiffusion | Experimental validation (VHH binders) | 4 disease-relevant epitopes targeted | [22] |
In silico success defined as AF2 structure with mean pAE <5, global backbone RMSD <2Ã…, and <1Ã… RMSD on scaffolded functional sites [20]
The performance advantages of LigandMPNN over traditional sequence design methods are particularly pronounced for metal-binding sites, where it more than doubles the sequence recovery rate of both Rosetta and ProteinMPNN [21]. This substantial improvement highlights the importance of explicitly modeling nonprotein atomic context when designing functional sites. The model's performance remains consistently superior across most proteins in validation datasets, with variations likely reflecting differences in crystal structure quality and native amino acid composition at binding sites [21].
RFdiffusion demonstrates remarkable capability in generating diverse protein topologies spanning alpha, beta, and mixed alpha-beta structures. The designs are not only structurally novel but also highly designable, as evidenced by AlphaFold2 and ESMFold predictions that closely match design models even for large proteins up to 600 residues [20]. Experimental characterization of several designed proteins revealed circular dichroism spectra consistent with design models and exceptional thermostability, confirming the practical utility of these computational designs [20].
Experimental Success in Therapeutic Antibody Design
Table 2: Experimentally validated antibody designs generated using RFdiffusion and ProteinMPNN
| Target Antigen | Antibody Format | Experimental Validation | Affinity (Kd) | Key Structural Validation |
|---|---|---|---|---|
| Influenza haemagglutinin | VHH (single-domain) | Yeast display, SPR, Cryo-EM | Initial: tens-hundreds nMMatured: single-digit nM | Cryo-EM confirms designed binding poseHigh-resolution structure verifies atomic accuracy of CDRs |
| Clostridium difficile toxin B (TcdB) | VHH (single-domain) | E. coli expression, SPR, Cryo-EM | Initial: tens-hundreds nM | Cryo-EM confirms designed binding pose |
| Respiratory syncytial virus (RSV) sites I & III | VHH (single-domain) | Yeast surface display | Not specified | Binding confirmation at specified sites |
| SARS-CoV-2 RBD | VHH (single-domain) | Yeast surface display | Not specified | Binding confirmation |
| IL-7Rα | VHH (single-domain) | E. coli expression, SPR | Initial: tens-hundreds nM | Binding confirmation |
| TcdB | scFv (single-chain) | Cryo-EM | Not specified | Cryo-EM confirms binding poseHigh-resolution data verifies all 6 CDR loops |
| PHOX2B peptide–MHC complex | scFv (single-chain) | Not specified | Not specified | Not specified |
The experimental success of AI-designed antibodies represents a landmark achievement in computational protein design. For therapeutic antibody development, the fine-tuned RFdiffusion model has enabled targeting of diverse disease-relevant epitopes across viral pathogens, bacterial toxins, and immune signaling molecules [22]. Although initial computational designs typically exhibit modest affinities in the tens to hundreds of nanomolar range, subsequent affinity maturation using systems like OrthoRep can improve binding to single-digit nanomolar levels while maintaining epitope specificity [22].
The structural validation of designed antibodies is particularly noteworthy. Cryo-electron microscopy confirmed that designed VHHs targeting influenza haemagglutinin and Clostridium difficile toxin B bind in precisely the intended poses [22]. Even more impressively, high-resolution structure determination verified atomic accuracy of the designed complementarity-determining regions—a level of precision that was previously unimaginable for de novo antibody design. For single-chain variable fragments (scFvs) targeting TcdB, structural analysis confirmed the accurate design of all six CDR loop conformations [22].
Protocol: De Novo Antibody Design Pipeline
Stage 1: Structure Generation with Fine-Tuned RFdiffusion
Materials and Reagents:
- Target protein structure (PDB format or AlphaFold2 prediction)
- Specified epitope residues on target
- Antibody framework structure (e.g., h-NbBcII10FGLA for VHHs) [22]
- Computing resources: High-performance GPU cluster
- Software: RFdiffusion fine-tuned for antibody design [22]
Procedure:
- Input Preparation: Prepare the target protein structure and identify epitope residues for targeting. Select an appropriate antibody framework—for single-domain antibodies, the humanized VHH framework h-NbBcII10FGLA has proven effective [22].
Framework Conditioning: Provide the framework structure as conditioning input to RFdiffusion using the template track. This encodes the framework as a 2D matrix of pairwise distances and dihedral angles, preserving its structural integrity while allowing CDR innovation [22].
Epitope Specification: Designate epitope residues using one-hot encoded "hotspot" features to direct CDR interactions toward the target site. This ensures the generated antibodies specifically engage the desired epitope [22].
Structure Generation: Execute RFdiffusion sampling starting from random noise. The self-conditioning mechanism will iteratively denoise the structure over multiple steps (typically 200), progressively refining CDR loops and antibody orientation relative to the target [20] [22].
Initial Filtering: Select generated antibody structures that maintain framework integrity while exhibiting diverse CDR conformations that complement the target epitope topology.
Stage 2: Sequence Design with LigandMPNN
Materials and Reagents:
- Generated antibody backbone structures from RFdiffusion
- Target structure with specified epitope
- Computing resources: Standard CPU or GPU
- Software: LigandMPNN [21]
Procedure:
- Input Configuration: Prepare the complex structure containing the generated antibody backbone and target protein. Ensure all nonprotein atoms (including key functional groups at the epitope) are included in the input structure.
Context Atom Selection: LigandMPNN automatically selects the 25 closest ligand atoms to each protein residue based on virtual Cβ and ligand atom distances. Verify appropriate context inclusion for CDR residues [21].
Sequence Design: Execute LigandMPNN using random autoregressive decoding to generate sequences. Sample multiple sequences (typically 8-10) per backbone to explore sequence space while maintaining structural compatibility [20] [21].
Sidechain Packing: Utilize LigandMPNN's integrated sidechain packing network to predict optimal sidechain conformations. The network predicts mixture distributions for chi angles and decodes them autoregressively to generate physically realistic rotamer placements [21].
Stage 3: Validation and Affinity Maturation
Materials and Reagents:
- Fine-tuned RoseTTAFold2 for antibody structure prediction [22]
- Rosetta software suite for ddG calculations
- Yeast display system for experimental screening [22]
- OrthoRep system for in vivo affinity maturation [22]
Procedure:
- Computational Validation:
- Use fine-tuned RF2 to predict structures of designed antibody-antigen complexes. This specialized RF2 variant incorporates target structure and epitope information during prediction to enhance accuracy [22].
- Filter designs where predicted structures closely match designed models (backbone RMSD <2Ã…) and exhibit high interface quality (Rosetta ddG).
Experimental Screening:
- Clone designed antibody sequences into yeast display vectors.
- Screen libraries for target binding using fluorescence-activated cell sorting.
- Isplicate and characterize binding clones using surface plasmon resonance to determine affinity and specificity [22].
Affinity Maturation:
- For initial binders with modest affinity (tens to hundreds of nM), implement affinity maturation using OrthoRep continuous evolution system [22].
- Screen matured libraries for improved binders while maintaining epitope specificity.
- Characterize optimized antibodies using structural methods (cryo-EM, X-ray crystallography) to verify binding mode preservation [22].
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Key research reagents and computational tools for AI-driven antibody design
| Category | Item | Specification/Format | Function in Workflow |
|---|---|---|---|
| Computational Models | RFdiffusion (antibody fine-tuned) | Python/PyTorch implementation | De novo generation of antibody structures with targeted CDRs |
| ProteinMPNN/LigandMPNN | Python implementation | Sequence design for structural scaffolds with ligand context | |
| Fine-tuned RoseTTAFold2 | Python implementation | Structure prediction validation for antibody-antigen complexes | |
| Antibody Frameworks | h-NbBcII10FGLA | VHH framework structure | Humanized single-domain antibody scaffold for VHH designs |
| Therapeutic scFv frameworks | Structure files | Stable single-chain variable fragment scaffolds | |
| Experimental Systems | Yeast display system | Saccharomyces cerevisiae | High-throughput screening of antibody binding |
| OrthoRep evolution system | Yeast-based | In vivo affinity maturation without throughput bottlenecks | |
| Surface plasmon resonance | Biacore or similar | Quantitative binding affinity measurements | |
| Structural Validation | Cryo-electron microscopy | Frozen-hydrated samples | Structural validation of antibody-antigen complexes |
| X-ray crystallography | Crystallized complexes | High-resolution structural verification | |
| 7-Hydroxy amoxapine-d8 | 7-Hydroxy amoxapine-d8, CAS:1189671-27-9, MF:C17H16ClN3O, MW:321.8 g/mol | Chemical Reagent | Bench Chemicals |
| Cycloguanil-d4hydrochloride | Cycloguanil-d4hydrochloride, MF:C11H15Cl2N5, MW:292.20 g/mol | Chemical Reagent | Bench Chemicals |
The integration of specialized computational tools with appropriate experimental systems creates a powerful pipeline for de novo antibody development. The fine-tuned RFdiffusion model enables targeted exploration of structural space, while LigandMPNN ensures optimal sequence compatibility with both the antibody structure and antigen interface [21] [22]. The inclusion of orthogonal validation methods, particularly the antibody-optimized RoseTTAFold2, provides crucial computational filtering before resource-intensive experimental testing [22].
For therapeutic development, the use of well-characterized humanized frameworks like h-NbBcII10FGLA reduces immunogenicity risks while providing stable structural scaffolds for CDR innovation [22]. Combined with high-throughput screening using yeast display and subsequent affinity maturation through OrthoRep, this toolkit enables rapid development of high-affinity therapeutic antibodies against virtually any target epitope [22].
Troubleshooting and Technical Considerations
Common Challenges and Solutions
Low Success Rates in Initial Design Generation:
- Problem: Insufficient diversity in generated CDR conformations or failure to engage target epitope.
- Solution: Adjust the hotspot residue weighting in RFdiffusion inputs to more strongly bias sampling toward the target epitope. Increase the number of design samples to explore broader structural space [22].
Poor Expression or Aggregation of Designed Antibodies:
- Problem: Designed sequences exhibit poor solubility or expression yields.
- Solution: Implement CAPE-Beam decoding strategy with ProteinMPNN to minimize immunogenicity risk and improve biophysical properties [23]. Incorporate aggregation prediction tools during sequence filtering.
Inaccurate Structure Predictions for Validation:
- Problem: Standard AlphaFold2 fails to accurately model antibody-antigen complexes, complicating computational validation [22].
- Solution: Utilize the fine-tuned RoseTTAFold2 specifically trained on antibody structures with epitope information. This specialized model significantly outperforms general-purpose structure prediction for antibody validation [22].
Moderate Affinity in Initial Designs:
- Problem: Initial designed antibodies show binding in the hundreds of nM range, insufficient for therapeutic applications.
- Solution: Implement orthogonal affinity maturation systems like OrthoRep that can efficiently explore sequence space around initial hits while maintaining epitope specificity [22].
Advanced Applications and Future Directions
The integration of RFdiffusion and ProteinMPNN has opened new frontiers in therapeutic protein design beyond conventional antibodies. Recent advances demonstrate the capability to design single-chain variable fragments (scFvs) with all six CDR loops designed de novo, as validated by cryo-EM structures showing atomic accuracy across all complementarity-determining regions [22]. This capability significantly expands the design space for therapeutic binders.
For enzyme design and small-molecule targeting, LigandMPNN enables precise engineering of binding pockets that explicitly consider the chemical properties of nonprotein components [21]. The method's ability to recover native-like sequences for metal-binding sites (77.5% recovery) far surpasses traditional computational approaches, highlighting its potential for designing metalloenzymes and catalytic antibodies [21].
The emerging paradigm of generative AI beyond natural diversity points toward even more ambitious applications. Protein language models trained on natural sequences can now generate synthetic proteins that maintain structural and functional coherence while exhibiting properties not found in nature [19]. As one researcher noted, "These AI models are trained with all known protein sequences on earth and learn the internal language or 'grammar' of proteins. Using this grammar, they are able to speak this language perfectly, generating completely new proteins that maintain structural and functional meaning" [19].
For the therapeutic antibody field specifically, these advances suggest a future where customized antibodies can be designed against virtually any target epitope with atomic-level precision, potentially revolutionizing treatment development for infectious diseases, cancer, and autoimmune disorders. The open availability of these tools on platforms like GitHub ensures widespread access, while commercial development through biotech companies like Xaira Therapeutics promises to translate these computational advances into clinical therapies [24].
The Engineer's Toolkit: Rational Design, Directed Evolution, and Display Technologies
Rational protein design represents a pivotal strategy in modern protein engineering, enabling the precise development of therapeutics with enhanced properties. This approach relies on the fundamental principle of establishing a structure-function relationship, frequently via molecular modeling techniques, to guide controllable amino acid sequence changes [25]. In the context of therapeutic antibody research, rational design provides a powerful methodology for improving affinity, specificity, stability, and reducing immunogenicity [8]. Unlike directed evolution, which introduces random mutations, rational design utilizes detailed structural knowledge—including amino acid sequences, three-dimensional structures, and mechanistic functional data—to inform targeted modifications via site-directed mutagenesis [25] [26]. The integration of advanced computational tools, such as artificial intelligence and machine learning for structure prediction, has significantly accelerated the design process, allowing for more efficient and predictive engineering of antibody-based therapeutics [8] [26].
Core Principles and Workflow
The overarching goal of rational protein design is to confer desired properties—such as enhanced thermostability, modified binding affinity, or increased solubility—through targeted genetic alterations. The general workflow is iterative, cycling through design, mutagenesis, expression, and characterization phases until the desired functional outcome is achieved [25]. The critical first step involves acquiring a high-resolution structure of the target protein, determined through methods like X-ray crystallography, cryo-electron microscopy (cryo-EM), or nuclear magnetic resonance (NMR) spectroscopy [27] [25] [28]. Computational-aided design (CAD) is then employed to analyze the structure and identify specific amino acid residues for mutation that are predicted to improve function [25]. This is followed by the practical implementation of these changes through site-directed mutagenesis to create the variant genes, which are then expressed and purified [25]. Finally, the newly engineered proteins undergo rigorous characterization to validate that the designed properties have been successfully implemented [25].
The following diagram illustrates the logical and experimental workflow of a rational design cycle, from structural analysis to functional validation.
Key Experimental Protocols
The DiRect Site-Directed Mutagenesis Protocol
The Dimer-mediated Reconstruction by PCR (DiRect) method is an advanced SDM technique designed for high efficiency and minimal background. It is particularly suited for rational design-based protein engineering (RDPE) as it eliminates the need for laborious DNA cloning and sequencing steps typically required by conventional methods [29]. The protocol achieves nearly perfect mutation rates by employing a series of three consecutive PCR reactions to incorporate the mutation and reconstruct the full expression construct [29].
Primer Design for DiRect
- Mutagenesis Primers: Both forward and reverse primers are designed with a 5' half comprising a 21-nucleotide (nt) complementary sequence containing the mutation site in the middle. The 3' half consists of a 20-nt sequence that is complementary to the template plasmid [29].
- Reconstruction Primers: Standard primers are designed to bind outside the mutated region to amplify the full-length plasmid.
PCR Reaction Setup
The following table summarizes the components and conditions for the three PCR stages.
Table 1: Reaction Setup for DiRect Mutagenesis Protocol
| Component/Condition | Mutagenesis PCR (MutPCR) | Reconstruction PCR with Outer Primer (RecPCR-out) | Reconstruction PCR with Inner Primer (RecPCR-in) |
|---|---|---|---|
| Template | 5 ng of plasmid (e.g., pBK) | 2 µL of MutPCR product (unpurified) | 2 µL of RecPCR-out product (unpurified) |
| Primers | 0.5 µM each mutagenesis primer | 0.5 µM each reconstruction outer primer | 0.5 µM each reconstruction inner primer |
| Polymerase | KOD -Plus- Mutagenesis Polymerase | KOD -Plus- Mutagenesis Polymerase | KOD -Plus- Mutagenesis Polymerase |
| Total Volume | 20 µL | 20 µL | 20 µL |
| Thermal Cycling | 30 cycles of:• 98°C for 10 sec• 55°C for 30 sec• 68°C for 45 sec | 30 cycles of:• 98°C for 10 sec• 55°C for 30 sec• 68°C for 2 min | 30 cycles of:• 98°C for 10 sec• 55°C for 30 sec• 68°C for 2 min |
| Alexidine-d10 | Alexidine-d10, MF:C26H56N10, MW:518.9 g/mol | Chemical Reagent | Bench Chemicals |
| Bisphenol A-13C12 | Bisphenol A-13C12, CAS:263261-65-0, MF:C15H16O2, MW:240.20 g/mol | Chemical Reagent | Bench Chemicals |
Post-PCR Processing
- Digestion: Following the RecPCR-in, the product is treated with DpnI to digest the methylated template plasmid.
- Transformation: The DpnI-treated product is directly used to transform competent E. coli cells.
- Verification: The mutation efficiency is exceptionally high. Colony PCR or sequencing of a few clones is sufficient to identify a correct mutant [29].
The technical workflow of the DiRect method, from initial amplification to final construct assembly, is depicted below.
Protocol for Dynamics Analysis via DEFMap
Understanding protein dynamics is crucial for rational design, as function is regulated by both tertiary structure and dynamic behavior [28]. The Dynamics Extraction From cryo-EM Map (DEFMap) method is a deep learning-based approach that quantitatively estimates local protein dynamics directly from cryo-EM density maps [28].
Procedure:
- Cryo-EM Map Preprocessing: Download the 3D cryo-EM density map from EMDB. Apply a 5 Ã… low-pass filter and unify the grid width to 1.5 Ã…/grid.
- Subvoxel Extraction: Extract local density data as subvoxels with grid lengths of 15 Ã…, centered on the position of existing heavy atoms in the corresponding atomic model (from PDB).
- Data Augmentation: Augment the dataset by rotating the subvoxels by 90° in the xy, xz, and yz planes.
- Model Application: Input the prepared subvoxels into the pre-trained 3D Convolutional Neural Network (3D CNN) model.
- Dynamics Prediction: The model outputs the predicted root-mean-square fluctuation (RMSF) values, which represent atomic fluctuations, providing quantitative dynamics information for the target protein.
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents and Materials for Rational Protein Design
| Item | Function/Benefit in Rational Design | Example Use Case |
|---|---|---|
| KOD -Plus- Mutagenesis Polymerase | High-fidelity DNA polymerase with proofreading activity, essential for accurate amplification in multi-step PCR mutagenesis. | DiRect mutagenesis protocol [29]. |
| Ile-δ1[13CH3]/Met-ε1[13CH3] (IM-labeling) | Isotopically labeled precursors for methyl-TROSY NMR; allow site-specific study of interactions and dynamics in large complexes. | Probing subunit interfaces in the 410 kDa RNA exosome complex [27]. |
| 4-trifluoromethyl-L-phenylalanine (tfmF) | Non-natural amino acid for 19F NMR spectroscopy; serves as a sensitive probe for local environment and dynamics, even in large assemblies. | Incorporation via amber codon suppression to study loop dynamics [27]. |
| TEMPO Spin Label | Paramagnetic label for Paramagnetic Relaxation Enhancement (PRE) NMR experiments; provides distance restraints. | Mapping proximity between subunits (e.g., Csl4 and Rrp41 entry loop) [27]. |
| DpnI Restriction Enzyme | Digests methylated parental DNA template, enriching for newly synthesized PCR product containing the desired mutation in bacterial transformation. | Post-PCR digestion in the DiRect protocol [29]. |
| Cell-Free Protein Synthesis (CF) System | Uses PCR-amplified linear DNA and cell extracts for rapid protein expression, omitting cloning and fermentation steps. | Coupling with DiRect for high-throughput screening of designed variants (DiRect-CF) [29]. |
| 4-(3,6-Dimethylhept-3-yl)phenol-13C6 | 4-(3,6-Dimethylhept-3-yl)phenol-13C6, CAS:1173020-38-6, MF:C15H24O, MW:226.31 g/mol | Chemical Reagent |
| Secalciferol-d6 | Secalciferol-d6, CAS:1440957-55-0, MF:C27H44O3, MW:422.7 g/mol | Chemical Reagent |
Data Analysis and Interpretation
Quantitative Outcomes of Rational Design Strategies
The success of rational design and associated protocols is measured by quantitative changes in protein properties. The following table compiles key performance data from cited studies.
Table 3: Quantitative Data from Protein Engineering Studies
| Method / System | Key Performance Metric | Result / Value | Application Context |
|---|---|---|---|
| DiRect-CF System [29] | Mutation Success Rate | Nearly 100% | Generation of thermostable 3-quinuclidinone reductase (RrQR) variants. |
| DiRect-CF System [29] | Total Process Time | ~1 day | From mutant gene design to protein analysis. |
| DEFMap [28] | Prediction Correlation (r) | 0.665 (±0.124) | Correlation between predicted and actual (MD-derived) dynamics (RMSF). |
| DEFMap [28] | Correlation from Raw Map Intensity | 0.459 (±0.179) | Baseline for DEFMap performance improvement. |
| Rational Design [8] | Marketed mAbs (FDA Approved) | 144 (as of Aug 2025) | Outcome of therapeutic antibody engineering. |
| Phage Display [8] | FDA-Approved Antibody Drugs | 16 | Platform for generating fully human antibodies. |
Application in Therapeutic Antibody Engineering
The principles of rational design are extensively applied in therapeutic antibody development. Key advancements include:
- Humanization: CDR grafting is used to convert murine antibodies into humanized versions, drastically reducing immunogenicity while retaining specificity. This approach produced the first approved humanized mAb, daclizumab, and the oncology blockbuster trastuzumab [8].
- Affinity Maturation: Structural knowledge of the antibody-antigen interface allows for targeted mutagenesis of CDR residues to enhance binding affinity.
- Effector Function Engineering: Rational design modifies the Fc region to tailor interactions with immune cells, optimizing mechanisms of action like Antibody-Dependent Cellular Cytotoxicity (ADCC) for specific therapeutic goals [8].
- Bispecific Antibodies (bsAbs): Rational design enables the engineering of bsAbs that can simultaneously bind two distinct antigens, for example, redirecting T cells to tumor cells [8]. The integration of AI and machine learning is now revolutionizing these processes, improving immunogenicity prediction and enabling more efficient optimization of candidate antibodies [8].
Directed evolution stands as a cornerstone of modern protein engineering, harnessing the principles of natural selection in a controlled laboratory setting to tailor biomolecules for specific, human-defined applications. This powerful, iterative process of genetic diversification and functional selection has been instrumental in advancing therapeutic antibody research, enabling the development of antibodies with enhanced affinity, specificity, and stability [30]. For researchers and drug development professionals, mastering these techniques is crucial for accelerating the discovery of next-generation biologics.
The fundamental cycle of directed evolution involves two critical steps: first, the creation of genetic diversity within a parent gene to generate a vast library of variants, and second, the high-throughput screening or selection of this library to identify individuals with improved functional properties [31] [30]. Among the various methods for library generation, error-prone PCR (epPCR) is a widely adopted technique for introducing random mutations, mimicking the spontaneous mutations that drive natural evolution. Successive rounds of mutagenesis and screening allow for the accumulation of beneficial mutations, leading to proteins with significantly optimized performance characteristics for therapeutic use [32] [30].
Key Methodologies in Directed Evolution
The success of a directed evolution campaign hinges on the strategic choice of methods for creating diversity and for identifying improved variants. The following sections detail the core techniques, with a specific focus on epPCR and common screening platforms used in antibody engineering.
Generating Diversity: Error-Prone PCR (epPCR)
Error-prone PCR (epPCR) is a robust method for introducing random point mutations across a gene sequence without requiring prior structural knowledge [30]. This technique is particularly valuable in the early stages of antibody engineering when the goal is to explore a wide sequence space to enhance properties like affinity or stability [32].
The core principle involves modifying standard PCR conditions to reduce the fidelity of the DNA polymerase, thereby increasing the error rate during DNA amplification [30]. This is achieved through several key adjustments:
- Polymerase Selection: Using a polymerase that lacks 3' to 5' proofreading activity, such as Taq polymerase.
- Metal Cofactor: Adding manganese ions (Mn²âº) to the reaction, which is a critical factor for promoting misincorporation of nucleotides.
- dNTP Imbalance: Creating unbalanced concentrations of the four deoxynucleotide triphosphates (dNTPs) to further encourage misincorporation [30].
The mutation rate can be finely tuned by adjusting the concentration of Mn²âº, with typical protocols aiming for 1–5 base substitutions per kilobase of DNA, resulting in an average of one or two amino acid changes per protein variant [30]. It is important to note that epPCR is not perfectly random; it exhibits a mutational bias favoring transition mutations (AG, CT) over transversion mutations, which limits the accessible amino acid substitutions at any given position to an average of 5.6 out of 19 possible alternatives [30].
Table 1: Key Components and Conditions for a Standard Error-Prone PCR Protocol
| Component | Function | Considerations for epPCR |
|---|---|---|
| Template DNA | Gene of interest (e.g., scFv, Fab) | Use high-purity, minimal amount to avoid wild-type carryover. |
| Primers | Flank the gene for amplification | Design to anneal to constant regions outside the variable domains. |
| Taq Polymerase | Amplifies DNA | Lacks proofreading; essential for incorporating errors. |
| MgClâ‚‚ | Essential polymerase cofactor | Concentration is often elevated (~7 mM) to reduce fidelity. |
| MnClâ‚‚ | Key fidelity reducer | Titrate (0.1-0.5 mM) to control mutation rate [30]. |
| dNTPs | Nucleotide building blocks | Use unbalanced concentrations (e.g., higher dATP, dTTP) to promote errors. |
| PCR Buffer | Provides optimal reaction conditions | Standard buffer is used, but Mg²⺠and Mn²⺠are added separately. |
Advanced Cloning Technique: Circular Polymerase Extension Cloning (CPEC)
Following epPCR, the mutated gene fragments must be cloned into an expression vector for screening. Traditional, restriction enzyme- and ligase-dependent cloning methods are inefficient and can lead to a significant loss of library diversity [33]. Circular Polymerase Extension Cloning (CPEC) presents a highly efficient alternative.
CPEC uses a high-fidelity DNA polymerase to join the insert (e.g., the mutated antibody gene) and the linearized vector. The method relies on primers designed with overlapping sequences for the vector ends. During the PCR-based reaction, the polymerase extends these overlapping regions, seamlessly assembling a circular, replication-competent plasmid [33]. Studies have demonstrated that CPEC accelerates library generation and yields a greater number of functional variants compared to traditional methods, thereby better preserving the diversity created by epPCR [33].
Screening and Selection Platforms
Screening is often the rate-limiting step in directed evolution. The choice of platform depends on the desired antibody property and the required throughput.
Cell Surface Display: This is a powerful selection (not just screening) technology that physically links the genotype (the antibody gene) to the phenotype (the displayed antibody protein) [34]. Common systems include:
- Yeast Surface Display: A eukaryotic system that facilitates proper folding and post-translational modification of antibody fragments. It is compatible with fluorescence-activated cell sorting (FACS) for high-throughput screening of binding affinity and specificity [32] [34].
- Phage Display: Antibody fragments are displayed on the surface of filamentous phage. Libraries are subjected to panning rounds against the target antigen to enrich for binders. Recent advances integrate next-generation sequencing (NGS) for deep analysis of library diversity and enrichment [34] [35].
- Mammalian Cell Display: Allows for the display of full-length IgG antibodies in a host system with the most relevant cellular machinery for complex proteins, enabling direct functional screening [34].
Microfluidic Screening: Technologies like droplet microfluidics enable the ultra-high-throughput screening of millions of variants by compartmentalizing individual cells and assays in picoliter droplets, dramatically accelerating the discovery process [34].
Growth-Coupled Selection: For certain enzymatic functions, antibody activity can be coupled to the survival of the host cell. This creates a direct selection pressure where only cells producing functional antibodies proliferate, automating the enrichment process [36].
Table 2: Comparison of Common Screening and Selection Platforms for Antibody Engineering
| Platform | Throughput | Key Advantage | Primary Application in Antibody Research |
|---|---|---|---|
| Yeast Surface Display | High (FACS) | Eukaryotic processing; quantitative FACS | Affinity maturation, stability engineering |
| Phage Display | High (Selection) | Extremely large library sizes >10^11 [35] | De novo antibody discovery, affinity enrichment |
| Mammalian Cell Display | Medium-High | Full-length IgG display; native secretion | Functional screening of therapeutic candidates |
| Droplet Microfluidics | Very High (>10â·/day) | Massive parallelization, single-cell analysis | Ultra-high-throughput screening of binding |
| Growth-Coupled Selection | Continuous | Automates selection; minimal intervention | Evolving enzymes for antibody-drug conjugates |
Experimental Protocols
Detailed Protocol: Error-Prone PCR for Antibody Gene Mutagenesis
This protocol is designed to introduce random mutations into the variable regions of an antibody gene (e.g., scFv) using epPCR.
Materials:
- Template DNA (e.g., plasmid containing the antibody gene to be evolved)
- Gene-specific forward and reverse primers
- Taq DNA Polymerase (without proofreading)
- 10X PCR Buffer (with MgClâ‚‚)
- MnClâ‚‚ solution (e.g., 10 mM)
- dNTP Mix (can be standard or unbalanced)
- Nuclease-free water
Procedure:
- Prepare Reaction Mix: Set up a 50 µL PCR reaction on ice:
- 10X PCR Buffer: 5 µL
- dNTP Mix (0.2 mM each): 1 µL
- Forward Primer (10 µM): 1 µL
- Reverse Primer (10 µM): 1 µL
- Template DNA (10-100 pg): 1 µL
- MnCl₂ (10 mM): 0.5 µL (final conc. 0.1 mM)
- Taq DNA Polymerase (5 U/µL): 0.5 µL
- Nuclease-free H₂O: to 50 µL
Run PCR: Place the tube in a thermal cycler and run the following program:
- Initial Denaturation: 94°C for 2 min
- 30 Cycles of:
- Denaturation: 94°C for 15 sec
- Annealing: 60°C for 30 sec
- Extension: 68°C for 1 min/kb of gene length
- Final Extension: 68°C for 5 min
- Hold: 4°C
Purify Product: Run the PCR product on an agarose gel, excise the correct band, and purify using a gel extraction kit. Quantify the DNA concentration.
Note: The mutation rate can be adjusted by varying the MnCl₂ concentration (0.1–0.5 mM) or by using a commercial error-prone PCR kit [30].
Detailed Protocol: Cloning epPCR Products with CPEC
This protocol describes the cloning of purified epPCR products into a linearized expression vector using CPEC.
Materials:
- Purified epPCR product (insert)
- Linearized plasmid vector (e.g., for yeast surface display)
- High-fidelity DNA polymerase (e.g., LA Taq)
- Corresponding buffer and dNTPs
Procedure:
- Primer Design: Design primers for the insert so that their 5' ends have 15-25 bp overlaps with the ends of the linearized vector.
- Amplify Insert with Overlaps: Perform a PCR with the purified epPCR product as the new template, using the overlapping primers and a high-fidelity polymerase to generate the final "mutant insert."
- CPEC Reaction: Set up a 20 µL reaction:
- Linearized Vector: 50-100 ng
- Mutant Insert: 100-200 ng (use a 2:1 molar ratio of insert:vector)
- 2X CPEC Buffer (or high-fidelity PCR buffer): 10 µL
- High-fidelity Polymerase: 1 µL
- Nuclease-free H₂O: to 20 µL
- Run CPEC Program:
- Initial Denaturation: 94°C for 2 min
- 10-15 Cycles:
- Denaturation: 94°C for 15 sec
- Annealing: 55-65°C (Tm of overlaps) for 30 sec
- Extension: 68°C for 1-2 min/kb of total vector + insert length
- Final Extension: 68°C for 10 min
- Hold: 4°C
- Transform: Directly transform 5 µL of the CPEC reaction into competent E. coli cells for plasmid propagation before proceeding to screening in the final host system (e.g., yeast) [33].
Workflow Visualization
The following diagram illustrates the complete iterative cycle of directed evolution for therapeutic antibody engineering.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents for Directed Evolution of Antibodies
| Reagent / Solution | Function / Application | Example Notes |
|---|---|---|
| GeneMorph II Random Mutagenesis Kit | Commercial epPCR | Provides optimized conditions for controlled mutation rates [33]. |
| Taq DNA Polymerase | Low-fidelity PCR | Essential for standard epPCR; lacks proofreading [30]. |
| Linearized Display Vector | Library cloning | Backbone for yeast (e.g., pYD1) or phage (e.g., pIII) display systems. |
| Electrocompetent E. coli | Library propagation | High-efficiency strains (e.g., TOP10) for maximum library diversity. |
| Fluorescently Labeled Antigen | FACS Screening | Crucial for isolating high-affinity binders from yeast display libraries. |
| Magnetic Beads (Streptavidin) | Phage Display Panning | For immobilizing biotinylated antigen during selection rounds [34]. |
| Next-Generation Sequencing (NGS) | Library Diversity Analysis | Deep sequencing to validate library quality and track variant enrichment [34]. |
| tert-Butyl 1H-indol-4-ylcarbamate | tert-Butyl 1H-indol-4-ylcarbamate, CAS:819850-13-0, MF:C13H16N2O2, MW:232.28 g/mol | Chemical Reagent |
| (Rac)-Folic acid-13C5,15N | (Rac)-Folic acid-13C5,15N, CAS:1207282-75-4, MF:C19H19N7O6, MW:446.36 g/mol | Chemical Reagent |
The structured application of error-prone PCR and advanced library screening methodologies provides a powerful framework for the directed evolution of therapeutic antibodies. The iterative cycle of creating genetic diversity and applying stringent, high-throughput selection enables researchers to navigate vast sequence landscapes and isolate variants with enhanced properties such as picomolar affinity, high specificity, and excellent developability [34] [35]. As these protocols continue to be integrated with automation, microfluidics, and next-generation sequencing, the pace of engineering novel antibody-based therapeutics will only accelerate, offering robust solutions to complex challenges in biomedicine [37] [34].
In the field of therapeutic antibody development, in vitro display technologies represent a cornerstone of protein engineering, enabling the high-throughput screening and optimization of antibody candidates. These technologies physically link a protein (the phenotype) to its genetic information (the genotype), a principle known as genotype-phenotype coupling [38] [39]. This coupling allows for the rapid selection of specific binders from vast diverse libraries. Among the most prominent platforms are phage, yeast, and mammalian display, each with distinct advantages tailored to different stages of the discovery pipeline. The choice of display system significantly impacts the quality, functionality, and developability of the resulting therapeutic leads. This application note provides a comparative analysis of these three key technologies, detailing their methodologies and applications within the context of modern antibody engineering for drug development.
Technology Comparison at a Glance
The table below summarizes the core characteristics of phage, yeast, and mammalian display technologies to guide platform selection.
Table 1: Comparative Overview of Key Antibody Display Technologies
| Feature | Phage Display | Yeast Display | Mammalian Display |
|---|---|---|---|
| Typical Library Size | 10^10 – 10^12 [38] [39] | 10^8 – 10^9 [38] [39] | Up to 10^7 – 10^9 [40] [41] |
| Common Antibody Formats | scFv, Fab, VHH/sdAb [40] [39] | scFv, Fab, full-length IgG [40] [38] | Full-length IgG, scFv [40] [41] |
| Display Host | Bacteriophage (M13) [39] | Yeast (S. cerevisiae) [40] | Mammalian Cells (e.g., CHO, HEK293) [41] |
| Key Selection Method | Biopanning [6] | Fluorescence-Activated Cell Sorting (FACS) [40] [38] | FACS, Magnetic-Activated Cell Sorting (MACS) [38] [41] |
| Primary Advantages | Very large library sizes; robust and cost-effective; well-established [40] [6] | Eukaryotic secretion and folding; direct FACS for multiparametric sorting [40] [38] | Native PTMs; full-length IgG display; superior developability prediction [38] [41] |
| Key Limitations | Displays fragments only; bacterial folding may not reflect mammalian context [38] [39] | Smaller library sizes; non-human glycosylation [38] [39] | Smaller library sizes (though improving); more complex and costly [40] [41] |
| Ideal for Hit Identification On | Soluble antigens, peptides, cell surfaces [39] | Soluble antigens, affinity maturation [40] | Membrane proteins in native conformation, complex antigens on cell surfaces [41] |
Detailed Methodologies and Workflows
Phage Display Protocol
Phage display is a well-established in vitro selection technique where antibody fragments are expressed as fusions to a coat protein on the surface of bacteriophages, most commonly the M13 filamentous phage [6] [39].
Key Reagents:
- Phagemid Vector: A plasmid containing the antibody gene fused to the phage protein gene (e.g., pIII), a bacterial origin of replication, and a phage origin [6] [39].
- Helper Phage: Provides all other proteins needed for phage assembly and infection. "Hyperphage" lacking gene 3 can be used to enhance antibody display valency [39].
- E. coli Strain: An F+ strain capable of being infected by filamentous phage (e.g., TG1, XL1-Blue) [6].
- Antigen: Immobilized on a solid surface (e.g., immunotube, magnetic beads) or presented on cells.
Protocol: Biopanning for Hit Identification
- Library Incubation: The phage antibody library is incubated with the immobilized target antigen to allow binding.
- Washing: Non-specific or weakly binding phages are removed through extensive washing with buffers, sometimes containing mild detergents to reduce background.
- Elution: Specifically bound phages are eluted, typically using an acidic buffer (e.g., glycine-HCl, pH 2.2) or a competitive elution with the soluble antigen.
- Amplification: Eluted phages are used to infect a log-phase culture of E. coli. Co-infection with helper phage (if using a phagemid system) enables the production of new phage particles for the next selection round.
- Iteration: Steps 1-4 are repeated for 2-4 rounds to enrich the pool for high-affinity binders. The success of enrichment is monitored by comparing the phage titer (colony-forming units) of the output eluate against a control [6].
- Screening: After the final round, individual clones are picked, and their binding specificity and affinity are characterized using ELISA, sequencing, and other binding assays [6].
The following workflow diagram illustrates the biopanning cycle:
Yeast Display Protocol
Yeast surface display leverages the eukaryotic quality control machinery of Saccharomyces cerevisiae to express and anchor antibodies to its cell wall, typically via the Aga1-Aga2 agglutinin system [40].
Key Reagents:
- Yeast Display Vector: Plasmid encoding the antibody gene (e.g., scFv) fused to Aga2p, and a selection marker for maintenance in yeast.
- Antigen: Purified and labeled with a fluorophore (e.g., biotinylated for detection with streptavidin-PE).
- Detection Antibodies: Antibodies against epitope tags (e.g., c-Myc, HA) fused to the antibody for detection of surface expression.
- FACS Aria or Similar: Fluorescence-activated cell sorter capable of high-throughput multiparametric analysis and sorting.
Protocol: FACS-Based Selection
- Induction: The yeast library is cultured under conditions that induce the expression of the antibody-Aga2p fusion.
- Labeling: Induced yeast cells are labeled with two distinct reagents:
- The target antigen, fluorescently labeled (e.g., Antigen-PE).
- A primary antibody against an epitope tag (e.g., anti-c-Myc), followed by a fluorescently labeled secondary antibody (e.g., FITC) to monitor total surface expression.
- FACS Analysis and Sorting: Labeled cells are analyzed by flow cytometry. Cells are gated based on their dual fluorescence:
- High Antigen Binding & High Expression: These cells, which represent the highest-affinity binders, are sorted into a collection tube.
- Recovery and Iteration: Sorted cells are grown in rich media to allow for recovery and expansion. The process is repeated for 2-3 rounds, with increasing stringency (e.g., reduced antigen concentration) to drive affinity maturation [40].
- Characterization: After sorting, plasmid DNA is isolated from the enriched population, and individual clones are sequenced and characterized for binding affinity.
The logic of the FACS gating strategy is central to the success of yeast display:
Mammalian Display Protocol
Mammalian display presents antibodies on the surface of mammalian cells (e.g., CHO, HEK293), enabling expression of full-length, natively folded and glycosylated IgGs in their natural context [38] [41].
Key Reagents:
- Lentiviral Vector: For highly efficient delivery and stable integration of the antibody gene library into the host cell genome.
- Engineered Mammalian Cell Line: Cells that may also be engineered to express a specific target membrane protein for self-labeling selections [41].
- Detection Reagents: Fluorescently conjugated anti-human Fc antibodies (for IgG display) or anti-tag antibodies (for scFv formats).
- MACS Columns & Beads: For initial enrichment (e.g., Anti-PE MicroBeads).
- FACS Aria: For high-precision, single-cell sorting based on complex phenotypic signatures.
Protocol: Self-Labeling Selection for Membrane Protein Targets
A powerful application of mammalian display is the selection of antibodies against membrane proteins in their native conformation, using a "self-labeling" system [41].
- Library Transduction: A lentiviral library encoding a diverse pool of antibodies (e.g., scFv-HA) is used to transduce a mammalian cell line (e.g., CHO) that has been engineered to express the target membrane protein (e.g., EpCAM).
- Self-Labeling: Post-transduction, cells co-express the target protein and secrete the antibody library. Cells producing antibodies that bind the membrane protein on their own surface become "self-labeled."
- MACS Enrichment: Self-labeled cells are incubated with an anti-HA-PE antibody. These cells are then captured using anti-PE magnetic beads and isolated via MACS. This step rapidly enriches for potential binders from a large pool [41].
- FRET-Based FACS: To precisely identify cells where the secreted antibody is bound to the intended target (and not other surface proteins), a FRET (Förster Resonance Energy Transfer)-based FACS assay can be employed. Cells are stained with two different fluorophores: one on the target protein (donor) and one on the secreted antibody (acceptor). A FRET signal only occurs when the two are in very close proximity, confirming specific interaction.
- Single-Cell Sorting and Recovery: FRET-positive single cells are sorted into 96-well plates. The viral DNA integrated into the host genome is then recovered via PCR, sequenced, and the antibody genes are cloned for recombinant expression and functional validation [41].
The self-labeling workflow for challenging membrane protein targets is illustrated below:
The Scientist's Toolkit: Essential Research Reagents
Successful execution of display technology campaigns relies on a suite of specialized reagents and tools. The following table details key solutions for researchers in this field.
Table 2: Essential Research Reagent Solutions for Display Technologies
| Reagent / Solution | Function & Application | Technology Platform |
|---|---|---|
| Phagemid Vectors & Helper Phage | Engineered plasmids and helper viruses for packaging and displaying antibody fragments on phage surface. | Phage Display [6] [39] |
| Yeast Display Vectors (Aga2p) | Plasmids for inducible, surface-anchored expression of antibody fragments in S. cerevisiae. | Yeast Display [40] |
| Lentiviral Packaging Systems | Systems for generating high-titer lentivirus to efficiently deliver antibody libraries into mammalian cells for stable expression. | Mammalian Display [41] |
| Biotinylated Antigens | Purified antigens conjugated to biotin, used with streptavidin-coated magnetic beads or fluorescent streptavidin for selection and detection. | All Platforms |
| Fluorophore-Conjugated Anti-Tag Antibodies | Antibodies against common epitope tags (HA, c-Myc, FLAG) for detecting and quantifying surface expression of displayed antibodies. | Yeast & Mammalian Display [41] |
| MACS MicroBeads & Columns | Magnetic beads conjugated to antibodies (e.g., anti-PE) and columns for rapid, high-throughput enrichment of labeled cells. | Mammalian Display (also Yeast) [41] |
| Fluorescent Cell Sorting Reagents | A wide range of fluorophores and kits optimized for labeling and staining cells for FACS analysis and sorting. | Yeast & Mammalian Display |
| 6-Amino-5-nitroso-2-thiouracil-13C,15N | 6-Amino-5-nitroso-2-thiouracil-13C,15N, MF:C4H4N4O2S, MW:174.15 g/mol | Chemical Reagent |
| DSP Crosslinker-d8 | DSP Crosslinker-d8, MF:C14H16N2O8S2, MW:412.5 g/mol | Chemical Reagent |
Phage, yeast, and mammalian display technologies offer a powerful, complementary toolkit for identifying and engineering therapeutic antibody hits. The choice of system involves a strategic balance between library diversity, the biological relevance of the display context, and screening throughput. Phage display remains unparalleled for accessing immense diversity at a lower cost, yeast display excels in quantitative, multiparametric affinity-based screening, and mammalian display is emerging as a superior platform for identifying functional antibodies against complex targets like native membrane proteins, with built-in advantages for predicting clinical developability. Integrating these technologies, such as by using phage for initial mining and mammalian display for late-stage optimization, represents a robust strategy for accelerating the discovery of next-generation biotherapeutics.
The field of therapeutic antibodies has evolved dramatically beyond conventional monoclonal antibodies, with protein engineering now enabling sophisticated multifunctional biologics. Three advanced formats—bispecific antibodies (BsAbs), antibody-drug conjugates (ADCs), and antibodies engineered for enhanced Fc-mediated effector functions—represent the forefront of this innovation. These engineered therapeutics leverage precise structural modifications to achieve enhanced targeting, potent cytotoxicity, and immune system engagement, offering new avenues for treating complex diseases, particularly in oncology.
BsAbs are synthetic molecules capable of simultaneously binding two distinct antigens or epitopes, enabling unique mechanisms such as immune cell recruitment and dual pathway inhibition [42] [43]. ADCs constitute a distinct class of targeted cytotoxics, combining the specificity of monoclonal antibodies with the potent killing power of chemical payloads, thereby functioning as "biological missiles" against cancer cells [44] [45]. Concurrently, engineering of the Fc region allows fine-tuning of effector functions like antibody-dependent cellular cytotoxicity (ADCC) and phagocytosis (ADCP), which are crucial for eliminating pathogen-infected or malignant cells [46] [47] [48]. This article provides detailed application notes and experimental protocols for developing and characterizing these advanced therapeutic formats within a protein engineering framework.
Bispecific Antibodies (BsAbs): Engineering Dual Targeting
Molecular Platforms and Engineering Strategies
BsAbs are fundamentally classified into two categories based on the presence or absence of an Fc region. IgG-like BsAbs (with Fc) benefit from longer half-life, improved solubility, and Fc-mediated effector functions like ADCC, CDC, and ADCP. Non-IgG-like BsAbs (without Fc), such as bispecific T-cell engagers (BiTEs), often exhibit superior tissue penetration and reduced Fc-mediated toxicity but have shorter half-lives [42] [43].
A primary challenge in BsAb production is the mispairing of heavy and light chains. Several engineered platforms address this:
- Knobs-into-Holes (KiH): This platform introduces sterically complementary mutations in the CH3 domain of the Fc region. A "knob" (T336Y substitution) is created in one heavy chain, while a "hole" (Y407T substitution) is formed in the other, promoting heterodimerization with an efficiency up to 95% [42].
- DuoBody: This controlled Fab-arm exchange (cFAE) platform relies on introducing complementary mutations (K409R and F405L) in the CH3 regions of two parental antibodies. When co-expressed, these antibodies undergo Fab-arm exchange to form the bispecific molecule [42].
- SEED (Strand-Exchange Engineered Domain): This platform uses alternating sequences from human IgA and IgG to create asymmetric AG and GA SEED CH3 domains that preferentially form heterodimers [42].
- ART-Ig (Asymmetric reengineering technology): Heterodimerization is promoted by introducing charged pairs (e.g., D360K/D403K in one chain and K402D/K419D in the other) in the Fc region. Emicizumab is a BsAb based on this platform [42].
Table 1: Key Engineering Platforms for IgG-like Bispecific Antibodies
| Platform | Core Engineering Principle | Key Feature | Example (Therapeutic) |
|---|---|---|---|
| Knobs-into-Holes | Steric complementarity in CH3 domain | High heterodimer yield (~95%) | Mosunetuzumab [43] |
| DuoBody | Controlled Fab-arm exchange | Dynamic recombination | JNJ-61186372 (EGFR/c-MET) [42] |
| SEED | CH3 domain from IgA/IgG hybrids | Asymmetric, complementary domains | Platform in preclinical development [42] |
| ART-Ig | Electrostatic steering in Fc | Charge-based chain pairing | Emicizumab (Factor IXa/X) [42] |
Application Notes: Functional Mechanisms and Applications
BsAbs exert their therapeutic effects through three primary mechanistic classes:
- Bridging Immune and Tumor Cells: T-cell engaging (TCE) BsAbs, such as those targeting CD3 and a tumor-associated antigen (e.g., CD19), create a cytolytic synapse between a T-cell and a cancer cell, leading to T-cell activation and tumor cell lysis independent of MHC recognition [43].
- Dual Signaling Pathway Inhibition: BsAbs like amivantamab (targeting EGFR and MET) simultaneously block multiple oncogenic signaling pathways, overcoming resistance to single-target inhibitors [43].
- Cofactor Mimicry: BsAbs can mimic natural cofactors to restore biological function. For instance, emicizumab bridges activated Factor IX and Factor X, restoring coagulation function in hemophilia A [42].
Experimental Protocol: Assessing T-Cell Engagement and Cytotoxicity
Objective: To evaluate the potency of a T-cell engaging BsAb in mediating the lysis of target tumor cells.
Materials:
- Purified BsAb (e.g., anti-CD3 x anti-tumor antigen)
- Effector cells: Human peripheral blood mononuclear cells (PBMCs) or purified T-cells
- Target cells: Tumor cell line expressing the target antigen
- Culture medium (e.g., RPMI-1640 with 10% FBS)
- â 96-well tissue culture plates
- Lactate dehydrogenase (LDH) release detection kit or flow cytometry with viability dyes (e.g., propidium iodide)
Method:
- Effector Cell Preparation: Isolate PBMCs from healthy donor blood using Ficoll density gradient centrifugation. Resuspend in culture medium.
- Target Cell Preparation: Harvest adherent tumor cells and resuspend in fresh medium. Count cells.
- Co-culture Setup: Seed target cells in a 96-well plate (e.g., 10,000 cells/well). Add effector cells at various effector-to-target (E:T) ratios (e.g., 10:1, 5:1, 1:1). Treat with a serial dilution of the BsAb. Include controls: effector cells alone, target cells alone, target cells with BsAb but no effector cells, and target cells with effector cells but no BsAb.
- Incubation: Incubate the plate for 24-48 hours at 37°C in a 5% CO₂ incubator.
- Cytotoxicity Measurement:
- LDH Release Assay: Centrifuge the plate. Transfer supernatant to a new plate and measure LDH activity according to the kit manufacturer's instructions. Calculate specific lysis using the formula:
Specific Lysis (%) = (Experimental - Effector Spontaneous - Target Spontaneous) / (Target Maximum - Target Spontaneous) * 100 - Flow Cytometry: Harvest co-cultured cells, stain with a viability dye, and analyze by flow cytometry. The percentage of dead target cells is determined by gating on the target cell population.
- LDH Release Assay: Centrifuge the plate. Transfer supernatant to a new plate and measure LDH activity according to the kit manufacturer's instructions. Calculate specific lysis using the formula:
- Data Analysis: Plot specific lysis (%) against BsAb concentration to generate a dose-response curve and determine the half-maximal effective concentration (ECâ‚…â‚€).
Diagram: Cytotoxicity Assay for T-cell Engagers
Antibody-Drug Conjugates (ADCs): Precision Delivery of Cytotoxics
Key Components and Engineering Evolution
ADCs are complex molecules composed of a monoclonal antibody, a cytotoxic payload, and a chemical linker. The evolution of ADCs is categorized into generations based on engineering improvements [45] [49]:
- First-Generation: Utilized murine antibodies and unstable linkers, leading to high immunogenicity and off-target toxicity (e.g., Gemtuzumab Ozogamicin).
- Second-Generation: Employed humanized antibodies and more stable linkers, improving target specificity and therapeutic index (e.g., Brentuximab Vedotin, Trastuzumab Emtansine).
- Third-Generation: Feature site-specific conjugation technologies and fully human antibodies, resulting in homogeneous Drug-to-Antibody Ratio (DAR) and reduced immunogenicity (e.g., Enfortumab Vedotin).
- Fourth-Generation: Incorporate novel payloads with high DAR and bystander effects, killing neighboring antigen-negative cells (e.g., Trastuzumab Deruxtecan).
Table 2: Core Components of Modern ADCs
| Component | Function & Ideal Properties | Common Examples |
|---|---|---|
| Antibody | Targets surface antigen. High specificity, internalization, humanized/low immunogenicity. | Trastuzumab (HER2), Cetuximab (EGFR) [44] [45] |
| Linker | Connects antibody and payload. Stable in circulation, cleavable in target cell (lysosome). | Cleavable: Valine-Citrulline (vc). Non-cleavable: SMCC [45] [49] |
| Payload | Kills the target cell. Highly potent (ICâ‚…â‚€ in pM-nM range), modifiable functional groups. | Tubulin inhibitors: MMAE, DM1. DNA damaging: Calicheamicin, Deruxtecan (DXd) [45] [49] [50] |
Application Notes: Mechanism of Action and Bystander Effect
The mechanism of ADC action involves: (1) binding to the target cell surface antigen, (2) internalization via receptor-mediated endocytosis, (3) trafficking to lysosomes, (4) degradation of the antibody and cleavage of the linker to release the payload, and (5) payload-induced cell death (e.g., via DNA damage or microtubule disruption) [45] [49].
A critical advancement is the bystander effect. Payloads with high membrane permeability (e.g., Deruxtecan), once released inside the target cell, can diffuse out and kill adjacent tumor cells, including those that are antigen-negative or heterogeneous. This is particularly effective against solid tumors [49].
Experimental Protocol: Evaluating ADC Internalization and Payload Release
Objective: To quantify the internalization efficiency of an ADC and the subsequent intracellular release of its cytotoxic payload.
Materials:
- ADC with a fluorescently labeled antibody (e.g., Alexa Fluor 488 conjugate) or a payload that can be detected via antibody.
- Target antigen-positive cell line.
- Control: Isotype control ADC or untransfected cell line.
- Flow cytometry buffer (PBS with 1% BSA).
- Acidic wash buffer (e.g., 50 mM glycine, 150 mM NaCl, pH 2.5-3.0) to remove surface-bound antibodies.
- Fixation/Permeabilization buffer kit.
- Flow cytometer or confocal microscopy system.
Method:
- Cell Seeding: Seed target cells in a 12-well plate and incubate overnight to achieve 70-90% confluence.
- ADC Binding: Chill cells and culture medium on ice. Add the fluorescent ADC to the cells and incubate on ice for 60-90 minutes. This allows binding without internalization.
- Internalization: Remove unbound ADC by washing with cold buffer. Add fresh, pre-warmed medium and shift the plate to a 37°C water bath or CO₂ incubator for different time points (e.g., 0, 15, 30, 60, 120 minutes).
- Acid Stripping: At each time point, return the plate to ice. Wash one set of wells with acidic buffer to strip surface-bound ADC. Wash a parallel set with neutral buffer (to measure total cell-associated fluorescence).
- Analysis:
- Flow Cytometry: Harvest cells (using enzyme-free dissociation buffer for adherent cells), fix, and analyze by flow cytometry. The internalized fraction is calculated as the acid-resistant fluorescence. For payload detection, permeabilize cells and stain with an antibody specific for the released payload.
- Confocal Microscopy: Fix cells after internalization, stain nuclei and endosomal/lysosomal markers (e.g., LAMP1), and image using a confocal microscope to visualize ADC trafficking.
- Data Analysis: Plot the mean fluorescence intensity (MFI) of the internalized fraction (acid-resistant) over time to generate an internalization kinetics curve.
Fc-Mediated Effector Functions: Engineering for Enhanced Immunity
Key Effector Functions and Their Mechanisms
The Fc region of an antibody mediates critical effector functions by engaging Fc receptors (FcRs) on immune cells or components of the complement system [46] [47] [48]. These functions are crucial for eliminating infected or malignant cells.
- Antibody-Dependent Cellular Cytotoxicity (ADCC): IgG antibodies bound to a target cell engage FcγRIII (CD16) on Natural Killer (NK) cells, triggering the release of perforin and granzymes that induce apoptosis in the target cell [46] [48].
- Antibody-Dependent Cellular Phagocytosis (ADCP): IgG-bound target cells are engulfed by macrophages via engagement of FcγRs [46] [47].
- Complement-Dependent Cytotoxicity (CDC): The C1q complex binds to clustered antibody Fc regions on the cell surface, initiating the complement cascade and forming a membrane attack complex (MAC) that lyses the target cell [46] [48].
Application Notes: Fc Engineering Strategies
Protein engineering enables the modulation of Fc-mediated effector functions to tailor therapeutic activity:
- Enhancing Effector Functions: For oncology applications, enhancing ADCC is desirable. This is achieved by engineering Fc domains for increased affinity to activating FcγRIIIa, such as through amino acid mutations (e.g., S298A/E333A/K334A) or by reducing the fucose content of the Fc-associated N-glycan (afucosylation), which can increase ADCC potency by up to 50-fold [46].
- Silencing Effector Functions: For some applications, like T-cell engaging BsAbs, Fc effector functions are undesirable as they can cause non-specific T-cell activation and cytokine release. Mutations like L234A/L235A (IgG1) or aglycosylation can ablate FcγR and C1q binding [43].
Experimental Protocol: Measuring Fc Effector Functions (ADCC/ADCP)
Objective: To assess the potency of an engineered antibody in inducing ADCC or ADCP.
Materials:
- Purified test antibody and controls (wild-type IgG, Fc-silenced mutant).
- Target cells (antigen-positive cell line).
- Effector cells: Peripheral blood mononuclear cells (PBMCs) for ADCC, or isolated monocytes/macrophages for ADCP.
- Flow cytometry buffer.
- For ADCC: Lactate dehydrogenase (LDH) release kit OR a flow-based assay using target cells pre-stained with a fluorescent dye (e.g., CFSE) and a viability dye (e.g., propidium iodide or 7-AAD).
- For ADCP: pHrodo-based phagocytosis assays are highly recommended, as pHrodo fluorescence increases dramatically in the acidic phagolysosome.
Method for Flow Cytometry-Based ADCC Assay:
- Label Target Cells: Stain target cells with CFSE (or similar) according to manufacturer's protocol.
- Setup Co-culture: Seed CFSE-labeled target cells in a U-bottom 96-well plate. Add effector cells (PBMCs) at a defined E:T ratio (e.g., 50:1). Add serial dilutions of the test antibody.
- Incubate: Incubate for 4-6 hours at 37°C, 5% CO₂.
- Stain for Viability: Add a viability dye (e.g., 7-AAD) to each well and incubate on ice for 15-30 minutes.
- Acquisition and Analysis: Analyze by flow cytometry. Gate on CFSE+ target cells. The percentage of ADCC is calculated as the proportion of CFSE+ cells that are also 7-AAD+ (dead). Plot this percentage against antibody concentration to determine potency.
Method for pHrodo-Based ADCP Assay:
- Label Target Cells: Label target cells with pHrodo dye according to the manufacturer's protocol.
- Setup Co-culture: Seed labeled target cells. Add monocyte-derived macrophages as effector cells. Add test antibodies.
- Incubate and Image: Incubate for 2-4 hours. Monitor phagocytosis in real-time using a fluorescence microscope or measure fluorescence intensity with a plate reader (excitation/emission ~560/585 nm).
- Analysis: Quantify the fluorescence intensity, which is directly proportional to the extent of phagocytosis.
Diagram: ADCC and ADCP Assay Workflows
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagent Solutions for Advanced Antibody Research
| Reagent / Solution | Function / Application | Key Considerations |
|---|---|---|
| Expression Vectors (e.g., Knobs-into-Holes) | Transient or stable expression of engineered BsAbs in mammalian cells (e.g., HEK293, CHO). | Ensure vectors encode the correct heavy and light chain pairs with appropriate selection markers [42]. |
| Site-Specific Conjugation Kits | For generating homogeneous ADCs with defined DAR. | Utilize technologies like engineered cysteines, unnatural amino acids, or enzymatic conjugation (e.g., transglutaminase) [45]. |
| Cytotoxic Payloads | The warhead of ADCs. | Select based on mechanism (tubulin inhibitor vs. DNA damaging), potency, and linkability. Novel payloads (e.g., PROTACs) are emerging [49] [50]. |
| Fc Receptor Binding Kits (SPR/BLI) | Quantify binding affinity of engineered Fc to human FcγRs (e.g., FcγRIIIa-V158). | Critical for characterizing Fc-engineered antibodies intended to enhance or silence effector functions [46]. |
| Human PBMCs / NK Cell Kits | Source of primary effector cells for in vitro functional assays (ADCC). | Isolate from fresh blood or use cryopreserved commercial sources. Maintain consistent E:T ratios [48]. |
| Antigen-Positive Cell Lines | Provide the target for functional assays. | Characterize antigen density (e.g., by flow cytometry) as it significantly impacts ADC/BsAb activity and effector function [44]. |
| Tridecane-d28 | Tridecane-d28, CAS:121578-12-9, MF:C13H28, MW:212.53 g/mol | Chemical Reagent |
The strategic engineering of bispecific formats, ADCs, and Fc-mediated functions represents a paradigm shift in therapeutic antibody development. By leveraging precise molecular platforms, optimized conjugation chemistries, and tailored Fc engineering, researchers can create next-generation biologics with enhanced efficacy and safety profiles. The application notes and detailed protocols provided herein offer a foundational framework for advancing research in this dynamic field, ultimately contributing to the development of more effective targeted therapies for cancer and other complex diseases.
Within the broader context of protein engineering for therapeutic antibodies, Fragment crystallizable (Fc) engineering has emerged as a powerful strategy to optimize the therapeutic profile of monoclonal antibodies (mAbs). The Fc domain mediates critical effector functions and determines antibody pharmacokinetics by engaging Fc gamma receptors (FcγRs) and the neonatal Fc receptor (FcRn) [51] [52]. Fc engineering enables the fine-tuning of these interactions, allowing researchers to enhance antibody-dependent cellular cytotoxicity (ADCC), antibody-dependent cellular phagocytosis (ADCP), complement-dependent cytotoxicity (CDC), serum half-life, and even blood-brain barrier (BBB) penetration [51] [53] [54]. This Application Note provides a structured overview of key Fc engineering strategies, summarizes quantitative data on engineered variants, and outlines detailed protocols for evaluating engineered antibodies in preclinical models, providing researchers with essential methodologies for advancing therapeutic antibody development.
Key Fc Engineering Strategies and Quantitative Comparisons
Enhancing Effector Functions via FcγR Engineering
Engineering the Fc domain to improve binding to activating Fcγ receptors can significantly enhance effector functions like ADCC and ADCP. Key approaches include introducing specific point mutations and generating asymmetrical Fc variants.
Table 1: Fc Variants for Enhanced FcγR Binding and Effector Functions
| Fc Variant | Amino Acid Substitutions | Key Functional Improvements | Experimental Evidence |
|---|---|---|---|
| DLE | S239D/A330L/I332E | Increased ADCC potency and serial killing by NK cells; Improved activation of NK cells [55]. | In vitro ADCC assays with AML cell lines; Single-cell TIMING analysis [55]. |
| DE (as in Tafasitamab) | S239D/I332E | Enhanced ADCC and ADCP; Clinically approved for DLBCL [54]. | Preclinical models of B-cell malignancies; Clinical trials [54]. |
| Asymmetrical Fc | Different mutations on each Fc chain | Enhanced ADCC; Improved selectivity for FcγRIIA over FcγRIIB; Superior CH2 domain stability [56]. | High-throughput screening (AlphaLISA); In vitro ADCC assays; Mouse xenograft models [56]. |
| Heterodimeric Hexamer | E345K (combined with S239D/I332E) | Enhanced CDC via promoted hexamerization while maintaining ADCC/ADCP [54]. | CDC and ADCC assays with isolated effector cells and whole blood [54]. |
Extending Serum Half-Life via FcRn Engineering
Optimizing the pH-dependent binding of the Fc domain to FcRn is a established strategy to prolong antibody half-life. The objective is to enhance binding in the acidic endosome (pH ~6.0) while ensuring rapid dissociation at neutral serum pH (pH 7.4) to avoid accelerated clearance.
Table 2: FcRn-Binding Variants for Improved Pharmacokinetics
| Fc Variant | Amino Acid Substitutions | Reported Half-Life Extension (vs. WT) | Key Characteristics |
|---|---|---|---|
| YTE | M252Y/S254T/T256E | 4.9-fold (Motavizumab) [57], 85 days in humans (vs. 26.5 days for WT) [58]. | Increased FcRn affinity at acidic and neutral pH; Used in clinically approved antibodies (e.g., Beyfortus) [58]. |
| LS | M428L/N434S | 8.7-fold (VRC01) [57], 71 days in humans (vs. 15 days for WT) [58]. | Enhanced FcRn binding; Used in Ultomiris and sotrovimab [58]. |
| YML | L309Y/Q311M/M428L | 6.1-fold (Trastuzumab) in hFcRn Tg mice [58]. | Superior FcRn association at acidic pH and rapid dissociation at neutral pH; Potent CDC activity [58]. |
| DHS | L309D/Q311H/N434S | Outperformed YTE and LS in hFcRn Tg mice [58]. | Fast dissociation at neutral pH is critical for prolonged half-life [58]. |
Enabling Blood-Brain Barrier Penetration
FcRn is expressed at the BBB and can be co-opted for receptor-mediated transcytosis of antibodies into the central nervous system (CNS). Engineering Fc for enhanced FcRn binding at the BBB can significantly increase brain uptake.
Table 3: Fc Engineering for Enhanced BBB Penetration
| Molecule/Target | Engineering Approach | Key Outcome | Experimental Validation |
|---|---|---|---|
| O4-YTE hIgG1 | YTE mutations (M252Y/S254T/T256E) | Widespread distribution throughout mouse brain parenchyma; FcRn-dependent transport [53]. | IHC in mouse brain; Knockout controls (Fcgr-/- mice); In vitro transcytosis assays [53]. |
ATVTfR |
Fc engineered to bind TfR | Enhanced brain exposure and unique parenchymal cell-type distribution in mice and cynomolgus monkeys [59]. | Whole-body tissue clearing/LSFM; FACS; scRNA-seq in mice; Biodistribution in primates [59]. |
ATVCD98hc |
Fc engineered to bind CD98hc | Enhanced brain exposure and distinct parenchymal cell-type distribution [59]. | Whole-body tissue clearing/LSFM; FACS; scRNA-seq [59]. |
Experimental Protocols
Protocol: Evaluating Pharmacokinetics of FcRn-Binding Variants in Tg32 Mice
This protocol describes how to assess the pharmacokinetic profile of Fc-engineered antibodies with enhanced FcRn binding using human FcRn transgenic (Tg32) mice, a critical step for predicting human PK [57].
Materials:
- Animals: Human FcRn transgenic mice (e.g., Tg32 strain).
- Test Articles: Fc-engineered mAbs and their wild-type (WT) counterparts.
- Reagents: Intravenous Immunoglobulin (IVIG), PBS, appropriate buffers.
Procedure:
- Formulate Antibodies: Prepare the Fc-engineered and WT mAbs in a sterile, injectable buffer such as PBS.
- Administer IVIG (Competitive Condition): To better mimic the competitive environment for FcRn binding in humans, inject a group of mice intravenously with a high dose of IVIG (e.g., 1000 mg/kg) shortly before antibody administration [57].
- Dose Mice: Intravenously administer the test antibodies (e.g., at 10 mg/kg) to the mice (with and without IVIG pre-treatment).
- Collect Plasma Samples: Serially collect blood samples at predetermined time points (e.g., 5 minutes, 6 hours, 1, 3, 7, 14, and 21 days post-injection) via a suitable method. Centrifuge blood samples to isolate plasma.
- Quantify Antibody Concentrations: Determine the concentration of the human therapeutic antibody in each plasma sample using a specific assay, such as an antigen-capture ELISA or surface plasmon resonance (SPR).
- Analyze Pharmacokinetic Data: Fit the plasma concentration-time data to a non-compartmental or a two-compartment model to estimate key PK parameters, including clearance (CL), volume of distribution in the central compartment (Vc), and half-life (t1/2). Compare the parameters of the Fc-engineered variant to the WT antibody, particularly under the IVIG competitive condition [57].
Protocol: Single-Cell Kinetic Analysis of ADCC Using TIMING
Time-lapse Imaging Microscopy in Nanowell Grids (TIMING) allows for high-resolution, quantitative analysis of the kinetics of ADCC at the single-cell level [55].
Materials:
- Effector Cells: Natural Killer (NK) cells, expanded and activated ex vivo.
- Target Cells: Antigen-positive tumor cell line.
- Test Antibodies: Fc-engineered and WT antibodies.
- Labeling Dyes: PKH26 (red) and PKH67 (green) fluorescent cell linkers.
- Viability Stain: Annexin V-AlexaFluor-647.
- Equipment: Nanowell array chip, automated live-cell imaging microscope.
Procedure:
- Label Cells:
- Label target cells with 1 µM red fluorescent dye (PKH26).
- Label NK effector cells with 1 µM green fluorescent dye (PKH67).
- Opsonize Target Cells: Incubate the labeled target cells with a sub-saturating concentration (e.g., 1 µg/mL) of the test antibody (Fc-engineered or WT) for 20 minutes at 4°C. Wash cells to remove unbound antibody.
- Load Nanowell Array:
- Sequentially load the opsonized target cells and effector cells onto the nanowell array at a specific E:T ratio.
- Immerse the array in phenol-red-free complete medium containing a viability stain like Annexin V-AlexaFluor-647.
- Acquire Time-Lapse Images: Place the chip in a live-cell imaging system and acquire images automatically every 7-10 minutes for 12-16 hours using a 20x objective.
- Analyze Single-Cell Data:
- Use image analysis software to segment and track individual cells over time.
- Quantify key kinetic parameters, including:
- Cytolytic Efficiency: The percentage of NK cells that become cytotoxic.
- Serial Killing: The number of target cells killed by a single NK cell.
- Kinetics of Apoptosis: The time from NK-target contact to target cell apoptosis (Annexin V positivity).
- Effector Cell Fate: Frequency of NK cell apoptosis after killing (activation-induced cell death).
Protocol: Assessing Brain Penetration of Fc-Engineered Antibodies
This protocol is used to evaluate the enhanced brain uptake and parenchymal distribution of antibodies engineered for improved FcRn binding or BBB transcytosis [53].
Materials:
- Animals: Wild-type mice and, if possible,
Fcgrt-/-(FcRn knockout) mice as a control. - Test Articles: Fc-engineered antibody (e.g., YTE variant) and WT control antibody.
- Reagents: Fixative (e.g., 4% PFA), primary antibody against human IgG, fluorescently-labeled secondary antibodies, IHC reagents.
Procedure:
- Administer Antibody: Intravenously inject mice with the test antibody (e.g., at 30 mg/kg) or a PBS vehicle control.
- Perfuse and Collect Tissues: At a predetermined terminal time point (e.g., 48 hours post-dose), deeply anesthetize the animals and transcardially perfuse them with ice-cold PBS to remove blood from the vasculature. Subsequently, harvest the brains and other tissues of interest.
- Process Brain Tissue: Fix brains in 4% PFA, followed by cryopreservation in sucrose. Embed tissue in OCT compound and section into thin slices (e.g., 20-40 µm) using a cryostat.
- Detect Human Antibody:
- For immunohistochemistry (IHC): Incubate brain sections with a primary antibody specific for human IgG, followed by a compatible detection system (e.g., fluorescently-labeled secondary antibody or enzyme-based chromogenic detection).
- For quantitative analysis: Homogenize brain tissue and measure human IgG concentration using a sensitive ELISA.
- Image and Analyze: Acquire high-resolution images of the stained brain sections using a fluorescence or brightfield microscope. Compare the distribution and intensity of the signal between the Fc-engineered antibody and the WT control.
- Key Analysis: Confirm FcRn-dependent transport by showing widespread parenchymal staining for the engineered variant (e.g., O4-YTE) that is absent in brains from
Fcgrt-/-mice [53].
- Key Analysis: Confirm FcRn-dependent transport by showing widespread parenchymal staining for the engineered variant (e.g., O4-YTE) that is absent in brains from
Visualization of Concepts and Workflows
Fc Engineering and Effector Functions
PK Evaluation Workflow in Tg32 Mice
The Scientist's Toolkit: Essential Research Reagents
Table 4: Key Reagents for Fc Engineering Research
| Reagent / Model | Function / Application | Key Consideration |
|---|---|---|
| Human FcRn Transgenic Mice (Tg32) | Preclinical PK prediction for human FcRn-binding antibodies [57]. | Co-injection with IVIG recommended to mimic human endogenous IgG competition [57]. |
FcRn Knockout Mice (Fcgrt-/-) |
Control model to confirm FcRn-dependent mechanisms in PK and BBB penetration studies [53]. | Essential for validating that enhanced brain delivery is specifically FcRn-mediated [53]. |
| IVIG (Intravenous Immunoglobulin) | Provides a source of competing endogenous IgG in PK studies in animal models [57]. | Critical for revealing the full PK benefit of FcRn-enhanced variants in non-competitive models like Tg32 mice [57]. |
| Cell Lines Stably Expressing FcRn & β2M | In vitro models for studying FcRn binding, internalization, and transcytosis [53]. | Overexpression of both subunits is often required for robust experimental readouts [53]. |
| TIMING (Time-lapse Imaging Microscopy in Nanowell Grids) | Single-cell kinetic analysis of ADCC, including serial killing and effector fate [55]. | Provides high-content, dynamic data beyond endpoint bulk cytotoxicity assays [55]. |
Navigating Developability Challenges: Immunogenicity, Stability, and Aggregation
Humanization and De-immunization Strategies to Mitigate Immune Responses
The development of therapeutic proteins, particularly monoclonal antibodies (mAbs), represents a cornerstone of modern biopharmaceuticals, with antibodies constituting half of all pharmaceutical sales [60]. A significant challenge in their development is immunogenicity, wherein a therapeutic protein triggers an unwanted immune response in patients. This can lead to the production of anti-drug antibodies (ADAs), resulting in altered pharmacokinetics, reduced efficacy, and potentially severe adverse effects like anaphylaxis or hypersensitivity reactions [61]. Immunogenicity stems from the immune system's ability to discriminate between self and non-self molecules. Therapeutic proteins derived from non-human sources, or even fully human proteins with engineered modifications, can be recognized as foreign [61] [62].
To address this, two complementary engineering philosophies have been developed: humanization and de-immunization. Humanization involves modifying protein therapeutics, especially those from non-human sources, to make them more similar to human counterparts. For antibodies, this often means transferring essential antigen-binding regions onto a human antibody framework [60]. De-immunization employs methods to remove or shield specific elements, known as T-cell and B-cell epitopes, that are recognized by the adaptive immune system, thereby reducing the potential for an immune response [61]. This application note details key strategies, protocols, and resources for implementing these approaches within a protein engineering workflow for therapeutic antibodies.
Background: Immunogenic Mechanisms of Therapeutic Proteins
The human immune response to protein therapeutics involves both innate and adaptive systems. The adaptive immune system, particularly T-cells and B-cells, plays the central role in the specific recognition of therapeutic proteins [61].
T-Cell Dependent Immunogenicity
The activation of T-cells is a critical step in initiating a persistent immune response. Antigen-presenting cells (APCs) endocytose the therapeutic protein, digest it into peptides, and load these peptides onto Major Histocompatibility Complex (MHC) molecules for presentation to T-cells.
- MHC Class I presents peptides derived from intracellular proteins to CD8+ cytotoxic T-cells.
- MHC Class II presents peptides from extracellular proteins to CD4+ helper T-cells, which are essential for providing help to B-cells for antibody production [61].
Even fully human sequence-derived antibodies can be immunogenic because their unique complementarity-determining regions (CDRs), especially the somatically mutated CDR3, are not subject to central tolerance and can be recognized as non-self [63]. For engineered proteins, junctions between fused human domains and point mutations create novel peptide sequences not found in the human proteome, which can be presented by MHC and activate T-cells [62].
Humanization Strategies for Therapeutic Antibodies
Antibody humanization has evolved from murine to chimeric, to humanized, and finally to fully human antibodies, with the goal of reducing immunogenicity while maintaining efficacy.
Table 1: Comparison of Key Antibody Humanization Techniques
| Technique | Description | Key Considerations | Example Therapeutics |
|---|---|---|---|
| Chimerization | Fusing variable regions of a non-human antibody with constant regions of a human IgG. | Retains ~33% non-human sequence; immunogenicity is reduced but not eliminated. | Abciximab [60] |
| CDR Grafting | Transferring the Complementarity-Determining Regions (CDRs) from a non-human antibody onto a human antibody framework. | Often requires additional "back-mutations" in the framework to preserve binding affinity and stability. | Trastuzumab [60] |
| SDR Grafting | Grafting only the Specificity-Determining Residues (a subset of CDRs critical for antigen binding) onto a human framework. | Further reduces non-human content but carries a higher risk of losing specificity and affinity. | - [60] |
| Resurfacing | Modifying only the surface-exposed residues in the variable regions to resemble human surface profiles. | Based on the assumption that buried residues are less likely to cause immunogenicity. | - [60] |
Protocol: Standard Workflow for CDR Grafting and Optimization
This protocol outlines the key steps for humanizing a murine monoclonal antibody via CDR grafting.
Sequence and Structural Analysis
- Obtain the amino acid sequence of the murine monoclonal antibody variable heavy (VH) and variable light (VL) chains.
- If available, use a 3D structure (e.g., from X-ray crystallography or homology modeling) to identify CDR and framework residues.
- Use the IMGT numbering scheme (via tools like ANARCI) for consistent alignment [64].
Human Template Selection
- Query the murine VH and VL sequences against a database of human germline V-genes (e.g., IMGT, VBASE).
- Select human acceptor frameworks with the highest sequence homology to the murine donor frameworks.
CDR Grafting
- Synthesize the humanized variable region sequences by grafting the murine CDR sequences (as defined by the Chothia or Kabat convention) onto the selected human framework regions.
Framework Back-Mutation Analysis
- Identify critical murine framework residues that influence CDR loop conformation or directly interact with the antigen (e.g., "Vernier" zone residues).
- Design constructs where these key murine residues are reintroduced ("back-mutated") into the human framework.
Construct Synthesis and Expression
- Clone the designed VH and VL sequences into expression vectors containing the desired human constant regions (e.g., IgG1).
- Transfect the plasmids into a mammalian expression system (e.g., HEK293 or CHO cells) and purify the antibody.
In Vitro Characterization
- Binding Affinity: Use surface plasmon resonance (SPR) or ELISA to confirm the humanized antibody retains affinity for its target antigen.
- Expression Yield & Biophysical Properties: Assess protein expression levels, solubility, and aggregation propensity via size-exclusion chromatography (SEC) and thermal stability assays.
Figure 1: CDR Grafting and Optimization Workflow. This diagram outlines the key experimental steps for humanizing a murine antibody, from initial analysis to a final candidate.
Computational De-immunization and Epitope Prediction
De-immunization strategies aim to directly eliminate T-cell and B-cell epitopes through rational design or machine learning.
T-Cell Epitope Prediction and Deletion
The most common de-immunization strategy involves identifying and mutating T-cell epitopes.
- Epitope Mapping: Computational tools predict peptide sequences within the therapeutic protein that can bind to MHC II molecules. These tools use algorithms ranging from position-specific scoring matrices (PSSMs) to advanced neural networks [61] [62].
- Deimmunization by Mutagenesis: Predicted epitopes are disrupted by introducing point mutations that reduce MHC binding affinity without compromising protein function. This can be a manual process or an automated, multi-objective optimization task [62].
Machine Learning for Antibody Humanization
Machine learning models trained on large-scale human antibody sequence datasets (e.g., Observed Antibody Space) can quantify the "humanness" of a sequence and guide humanization.
- Humanness Scoring: Models like Hu-mAb use random forest classifiers to distinguish human from non-human sequences. The resulting humanness score correlates negatively with experimental immunogenicity [64].
- Computational Humanization: Tools like Hu-mAb can systematically suggest mutations to a non-human input sequence to increase its humanness, effectively automating the initial design phase of humanization [64].
Table 2: Quantitative Metrics for De-immunization and Humanization
| Method | Metric | Typical Benchmark/Value | Interpretation |
|---|---|---|---|
| T-cell Epitope Prediction | Number of predicted MHC-II binders | Reduction from native to deimmunized variant | Fewer binders indicates lower T-cell activation potential. |
| Humanness Scoring (Hu-mAb) | Classification score (Human vs Non-human) | AUC: >0.99 [64] | Higher score indicates sequence is more "human-like". |
| Immunogenicity Correlation | Correlation between humanness score and immunogenicity | Negative relationship observed [64] | Higher humanness scores linked to lower immunogenicity. |
| B-cell Epitope Prediction | Spatial clustering of surface residues | N/A [61] | Identifies conformational B-cell epitopes for shielding. |
Protocol: A Machine-Guided Dual-Objective Engineering Workflow
This protocol describes a integrated computational and experimental pipeline for simultaneously optimizing protein function and deimmunization, applicable to novel scaffolds like zinc-finger proteins [62].
Generate Candidate Sequences
- Create a library of candidate protein sequences through:
- Domain Fusion: For fusion proteins, generate alternative combinations of protein domains.
- Directed Evolution/Mutagenesis: For single domains, introduce functional point mutations.
- Create a library of candidate protein sequences through:
In Silico Immunogenicity Screening
- For each candidate sequence, computationally digest the protein into peptides (e.g., 15-mers overlapping by 10).
- Use MHC-II prediction algorithms (e.g., NetMHCIIpan) to predict the binding affinity of each peptide against a panel of common HLA alleles.
- Calculate an aggregate immunogenicity score based on the number and strength of predicted MHC binders.
Multi-Objective Optimization
- Use a Pareto-optimization approach to identify candidate sequences that balance high predicted functionality (e.g., DNA-binding energy for transcription factors) with low predicted immunogenicity.
Construct Design and Experimental Validation
- Synthesize and clone the top-ranking deimmunized variants.
- Validate function using relevant assays (e.g., transcriptional activation for a synthetic transcription factor).
- Experimentally assess immunogenicity potential using in vitro T-cell activation assays (e.g., with human peripheral blood mononuclear cells - PBMCs).
Figure 2: Machine-Guided Dual-Objective Engineering. This workflow integrates immunogenicity prediction directly into the protein engineering cycle to simultaneously optimize for function and low immunogenicity.
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Resources for Humanization and De-immunization Research
| Reagent / Resource | Type | Function and Application | Examples / Notes |
|---|---|---|---|
| NSG Mouse Platform | In vivo model | Highly immunodeficient mouse strain enabling engraftment with human hematopoietic stem cells (HSCs) or peripheral blood mononuclear cells (PBMCs) to create "immune-humanized" models for preclinical testing [65]. | Useful for assessing immunogenicity and efficacy of therapeutics in a model with a human immune system. |
| ANARCI Software | Computational tool | Assigns International ImMunoGeneTics (IMGT) numbering to antibody sequences, ensuring consistent alignment for humanization and analysis [64]. | Critical first step for template selection and CDR definition. |
| Hu-mAb Web Server | Computational tool | Provides humanness scoring and suggests mutations for humanizing antibody variable domain sequences using machine learning models [64]. | Freely available at opig.stats.ox.ac.uk/webapps/humab. |
| OAS (Observed Antibody Space) | Database | A large, publicly available database of antibody sequences used for training machine learning models and comparative analysis [64]. | Contains nearly 2 billion redundant sequences. |
| ImmPort/ImmuneSpace | Data repository | Public repository of human immunology data, including systems vaccinology studies. Enables re-analysis of immune signatures [66]. | Useful for understanding correlates of immune responses. |
| Thera-SAbDab | Database | A database of therapeutic antibody sequences, structures, and metadata. Serves as a reference for approved and clinical-stage therapeutics [64]. | Useful for benchmarking and comparative analysis. |
The mitigation of immunogenicity is a critical hurdle in the development of effective biotherapeutics. A multi-faceted approach combining well-established techniques like CDR grafting with cutting-edge computational deimmunization strategies is essential. The field is moving towards integrated, machine-guided workflows that simultaneously optimize for therapeutic function and low immunogenicity from the earliest design stages. While fully human antibodies remain the gold standard, they do not guarantee a lack of immunogenicity, underscoring the need for continued innovation in deimmunization protocols. The tools and strategies outlined here provide a roadmap for researchers to engineer safer and more effective protein therapeutics.
The development of therapeutic antibodies presents a significant challenge: candidate molecules with excellent target affinity often exhibit poor biophysical properties, such as low solubility and inadequate chemical stability. These shortcomings can impede manufacturing, compromise formulation, reduce efficacy, and increase immunogenicity risk. As therapeutic applications advance toward high-concentration subcutaneous formulations and more complex modalities like antibody-drug conjugates (ADCs), the imperative for robust optimization strategies intensifies. This Application Note details integrated computational and experimental protocols for enhancing the solubility and conformational stability of therapeutic antibodies, providing a structured framework for researchers in protein engineering and drug development.
Computational Design for Stability and Solubility
Computational approaches enable the rational design of antibody variants with enhanced properties, significantly reducing experimental timelines and resource consumption.
Automated Pipeline for Multi-Trait Optimization
Advanced algorithms now facilitate the simultaneous optimization of conflicting traits, such as stability and solubility. The process resembles a "molecular Rubik's cube," where improving one property must not detrimentally impact others [67].
Key Algorithm Components:
- Input Requirements: Native structure or high-quality structural model of the target antibody.
- Phylogenetic Analysis: Uses multiple sequence alignments (MSA) to create a position-specific scoring matrix (PSSM), identifying evolutionarily tolerated mutations.
- Solubility Prediction: Leverages the CamSol method to predict changes in intrinsic solubility upon mutation and identify surface-exposed aggregation hotspots.
- Stability Prediction: Utilizes the FoldX energy function to calculate the thermodynamic impact of mutations on conformational stability.
Integrating phylogenetic filters dramatically reduces false positive predictions of stability, with the false discovery rate decreasing from approximately 26% to 15% [67]. The following workflow diagram illustrates the automated computational pipeline:
Protocol: Implementing the Computational Design Pipeline
Procedure:
- Structure Preparation: Obtain a high-resolution crystal structure or generate a reliable homology model of the antibody variable domain. Ensure the structure is properly protonated.
- Generate MSA: Using the antibody sequence as query, search protein databases (e.g., UniRef, OAS) for homologous sequences. For immunoglobulin variable domains, employ specialized tools that account for their modular nature.
- Construct PSSM: Calculate position-specific frequencies from the MSA. Compute log-likelihood scores for all possible mutations at each position.
- Run CamSol Analysis: Submit the wild-type structure to CamSol to identify surface patches with low solubility propensity and pinpoint specific aggregation-prone residues.
- Perform FoldX Stability Scan: Use FoldX to perform a systematic scan, calculating the ΔΔG of folding for all possible point mutations in the variable domain.
- Apply Joint Filtering:
- Retain mutations predicted by CamSol to improve solubility.
- Retain mutations predicted by FoldX to be stabilizing (ΔΔG < 0).
- Apply phylogenetic filter: prioritize mutations with positive Δlog-likelihood (observed more frequently in nature than wild-type residue).
- Combine Mutations: Select 3-5 top-ranking mutations for combinatorial library design, avoiding adjacent residues in the structure to minimize epistatic effects.
Technical Notes: The entire process is available through an automated webserver (www-cohsoftware.ch.cam.ac.uk) or can be implemented locally using published protocols [67].
Experimental Validation and Optimization
Computational designs require experimental validation to confirm improved properties while maintaining antigen binding.
Solubility and Stability Measurement Techniques
Multiple analytical techniques are employed to characterize biophysical properties:
Table 1: Analytical Techniques for Assessing Antibody Stability and Solubility
| Technique | Key Application | Sample Consumption | Throughput | Key Advantage | Key Limitation |
|---|---|---|---|---|---|
| Size Exclusion Chromatography with Multi-Angle Light Scattering (SEC-MALS) | Quantifying monomeric purity and aggregate levels [68] | High (µg per injection) | Moderate (30-60 min/sample) | Industry gold standard; provides molecular weight | Non-specific column interactions can skew results |
| Mass Photometry | Rapid aggregation screening under native conditions [68] | Very Low (ng per measurement) | High (minutes/sample) | Measures individual molecule mass; no separation needed | Optimal at nanomolar concentrations |
| Dynamic Light Scattering (DLS) | Initial size distribution assessment and aggregation screening [68] | Low | High | Rapid and easy to use; minimal sample prep | Low resolution; biased by large particles |
| Differential Scanning Calorimetry (DSC) | Measuring thermal unfolding transition midpoint (Tm) | Moderate | Low | Direct measurement of conformational thermal stability | Does not measure kinetic stability |
| Forced Degradation Studies | Assessing physicochemical stability under stress conditions [68] | Variable | Variable | Predicts long-term stability; guides formulation | Requires multiple analytical techniques for characterization |
Case Study: Enhancing a Catalytic Antibody Domain
A combined computational/experimental approach successfully enhanced the properties of the UA15-L variable domain, a catalytic antibody with poor thermostability and solubility [69].
Experimental Workflow:
- Identify Destabilizing Regions: Molecular dynamics simulations identified protein unfolding hotspots.
- Engineer Stabilizing Disulfide Bond: A disulfide bond was designed within an unfolding hotspot to restrict backbone flexibility and increase thermostability.
- Optimize Surface Solubility: Computational methods guided the introduction of polar or charged residues onto the protein surface to enhance solubility.
- Validate Designs: Combined mutations resulted in variants with improved expression, thermostability, solubility, and retained catalytic activity at elevated temperatures [69].
The following workflow summarizes the key experimental steps for validating computationally designed variants:
Protocol: Forced Degradation Study for Chemical Stability
Forced degradation studies evaluate an antibody's stability under stressors encountered during manufacturing, storage, and administration [68].
Materials:
- Purified antibody (>0.5 mg/mL in formulation buffer)
- Stressor solutions: 0.1 M Citrate (pH 3.0), 0.1 M Borate (pH 9.0), 3% Hydrogen Peroxide, 20 mM Histidine buffer (pH 6.0)
- Microcentrifuge tubes and thermomixer
- Mass photometer or SEC-MALS system
Procedure:
- Sample Preparation: Aliquot 50 µg of antibody into separate microcentrifuge tubes.
- Apply Stresses:
- Acidic Stress: Incubate with 0.1 M Citrate, pH 3.0, for 2 hours at 25°C.
- Basic Stress: Incubate with 0.1 M Borate, pH 9.0, for 2 hours at 25°C.
- Oxidative Stress: Incubate with 3% H₂O₂ for 2 hours at 25°C.
- Control: Incubate with 20 mM Histidine, pH 6.0, for 2 hours at 25°C.
- Neutralization: After stress incubation, return all samples to formulation buffer via dialysis or buffer exchange.
- Analysis: Analyze all samples and the unstressed control using mass photometry or SEC-MALS to quantify monomer loss and aggregate formation.
Interpretation: Compare the percentage of monomers and aggregates in stressed samples versus the control. Leads with less than 5% additional aggregation under most stress conditions generally exhibit superior developability [68].
Research Reagent Solutions
Successful optimization relies on specific reagents and platforms. The following table details key solutions used in the featured studies.
Table 2: Essential Research Reagents and Platforms for Antibody Optimization
| Reagent/Platform | Specific Example | Function in Optimization |
|---|---|---|
| Protein A Resin | MabSelect SuRe LX, MabSelect PrismA [70] | Affinity capture during purification; novel mild elution strategies can protect stability. |
| Stability Prediction Software | FoldX Algorithm [67] | Predicts changes in conformational stability (ΔΔG) upon mutation. |
| Solubility Prediction Software | CamSol Method [67] | Predicts intrinsic solubility and identifies aggregation-prone regions. |
| Mass Photometry System | Refeyn Mass Photometer [68] | Rapid, low-sample consumption measurement of aggregate levels under native conditions. |
| SEC-MALS System | Agilent HPLC with BioCore SEC-300 column [70] [68] | Industry-standard quantification of monomeric purity and aggregate molecular weight. |
| Amino Acid Elution Buffers | Leucine, Glycine, Serine buffers [70] | Enables milder elution from Protein A columns, minimizing acid-induced aggregation. |
The strategic optimization of solubility and chemical stability is a critical component of therapeutic antibody development. By integrating robust computational design with rigorous experimental validation, researchers can systematically overcome developability challenges. The automated computational pipeline for simultaneous stability and solubility optimization, coupled with analytical techniques like mass photometry and SEC-MALS for high-quality characterization, provides a powerful toolkit for advancing candidate molecules. Implementing these detailed protocols enables the engineering of antibodies with superior manufacturability, stability, and safety profiles, accelerating their path to clinical success.
For therapeutic monoclonal antibodies (mAbs), controlling aggregation is a critical quality attribute essential for ensuring drug safety and efficacy. Protein aggregation can occur during manufacturing, storage, and administration due to various stressors, potentially altering the immunomodulatory profile of the final product [71]. The Fragment crystallizable (Fc) region of IgG antibodies mediates effector functions by binding to Fc gamma receptors (FcγRs) on immune cells, initiating processes such as antibody-dependent cellular cytotoxicity (ADCC) and antibody-dependent cellular phagocytosis (ADCP) [71] [72]. This application note examines how antibody aggregates impact FcγR binding and downstream functional activity, providing detailed protocols for forced degradation studies, aggregate characterization, and binding affinity assessments to support robust therapeutic antibody development.
Key Findings: Aggregate-Mediated Modulation of FcγR Engagement
Recent investigations demonstrate that antibody aggregation significantly alters FcγR binding profiles, with implications for both analytical characterization and biological activity. A 2025 study systematically evaluated an effector-competent IgG1 (mAb1) subjected to forced degradation, revealing format-dependent effects on receptor binding [71].
Table 1: Impact of mAb1 Aggregation on FcγR Binding Across Assay Formats
| Fcγ Receptor | Avidity-Based SPR | Antibody-Down SPR | Solution Binding | Cell-Based Reporter Activity |
|---|---|---|---|---|
| FcγRIIa | Substantially Increased [71] | No Increase [71] | Increased [71] | Slight Increase [71] |
| FcγRIIIa | Increased [71] | Not Reported | Increased [71] | Decreased [71] |
| All FcγRs | Increased [73] [74] | Not Applicable | Increased [71] | Variable by Receptor [71] |
The observed effects are highly dependent on assay configuration. In avidity-based surface plasmon resonance (SPR) formats where FcγRs are immobilized on the chip surface, aggregated fractions showed dramatically increased binding across all FcγRs, with FcγRIIa particularly affected [71]. However, when measured using an antibody-down SPR format less susceptible to avidity effects, FcγRIIa binding showed no increase with aggregation [71]. This distinction highlights how aggregate-driven binding enhancements often reflect multivalent avidity effects rather than true affinity increases.
Functionally, these binding changes manifest differently across effector mechanisms. While FcγRIIa-mediated reporter activity slightly increased with aggregates, FcγRIIIa activity decreased, potentially due to altered glycosylation patterns in the aggregated material [71]. These findings underscore the critical need to monitor and control aggregation during manufacturing and formulation to ensure consistent therapeutic performance [73] [74] [71].
Experimental Protocols
Forced Degradation and Aggregate Generation
Purpose: To intentionally generate antibody aggregates under controlled stress conditions for evaluating FcγR binding alterations.
Materials:
- Purified therapeutic antibody (e.g., mAb1, IgG1) [71]
- Histidine buffer with NaCl, sucrose, and polysorbate 80, pH 6.0 [71]
- Hydrogen peroxide solution (1-10 ppm) [71]
- Iron chloride (0.5-20 ppm) and copper sulfate (20 ppm) solutions [71]
- 200 mM glucose solution [71]
- Borosilicate glass vials (20 mL) [71]
- Temperature-controlled incubation chamber
Procedure:
- Prepare antibody samples at 35 mg/mL in histidine buffer (pH 6.0) containing excipients.
- Aliquot samples into suitable glass containers and apply stress conditions:
- Oxidative Stress: Add hydrogen peroxide to final concentrations of 1 ppm and 10 ppm.
- Metal-Catalyzed Oxidation: Add iron chloride (0.5 ppm, 20 ppm) or copper sulfate (20 ppm).
- Glycation Stress: Add glucose to 200 mM final concentration.
- pH Stress: Adjust samples ±2 pH units from target pH 6.0.
- Incubate samples at 40°C for 28 days [71].
- Include dark controls by wrapping borosilicate glass vials with metal foil.
- For photostability assessment, expose samples to visible light at 1.2 million lux hours and 200 W·h/m² UV light at 20°C per ICH Q1B guidelines [71].
Aggregate Fractionation and Characterization
Purpose: To separate and quantify antibody aggregates generated through forced degradation.
Materials:
- Size Exclusion Chromatography (SEC) system (e.g., Agilent HPLC) [71]
- TSK-Gel G3000SWxl column (7.8 × 300 mm, 5 µm particles) [71]
- Mobile phase: 50 mM NaHâ‚‚POâ‚„, 300 mM NaCl, pH 7.0 [71]
- Fraction collection system
Procedure:
- Dilute stressed antibody samples to 1 mg/mL with mobile phase buffer.
- Perform SEC-HPLC with isocratic elution using the following parameters:
- Flow rate: 0.5 mL/min
- Column temperature: 25°C
- Injection volume: 10 µL
- Run time: 30 minutes
- Detection: UV at 214 nm [71]
- Collect fractions corresponding to monomeric and aggregated species based on retention time.
- Calculate percent aggregates as the sum of all peak areas eluting prior to the monomer peak [71].
- Confirm aggregate size ranges using complementary techniques such as analytical ultracentrifugation or light scattering as needed [71].
FcγR Binding Assessment by Surface Plasmon Resonance
Purpose: To quantify binding interactions between antibody aggregates and Fcγ receptors using multiple SPR formats.
Materials:
- SPR instrument (e.g., Biacore)
- CMS sensor chips
- Recombinant human Fcγ receptors (FcγRI, FcγRIIa, FcγRIIb, FcγRIIIa)
- Running buffer: HBS-EP (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.05% surfactant P20, pH 7.4)
- Amine coupling kit (if immobilizing)
Procedure for Avidity-Based Format (FcγR Immobilized):
- Immobilize FcγRs on CMS sensor chip using standard amine coupling chemistry to achieve approximately 500-1000 response units (RU).
- Dilute antibody fractions (monomer and aggregates) in running buffer and inject over FcγR surfaces at 30 µL/min for 3-minute association followed by 5-minute dissociation.
- Regenerate surface with 10 mM glycine, pH 2.0 between cycles.
- Reference subtract responses from control flow cell.
- Analyze binding responses during association phase to compare monomer vs. aggregate binding [71].
Procedure for Antibody-Down Format (Antibody Immobilized):
- Immobilize anti-human Fc antibody to capture test antibodies on sensor chip surface.
- Capture monomer or aggregate fractions to achieve consistent density (~50 RU).
- Inject FcγR analytes at multiple concentrations in single-cycle kinetics format.
- Measure binding responses to calculate affinity constants independent of avidity effects [71].
Cell-Based FcγR Reporter Assays
Purpose: To evaluate functional consequences of antibody aggregation on FcγR-mediated signaling.
Materials:
- FcγRIIa and FcγRIIIa reporter cell lines (engineered Jurkat cells expressing NF-κB-GFP reporter)
- Cell culture medium appropriate for reporter cells
- Flow cytometer or plate reader for GFP detection
- Positive control antibody (rituximab for FcγRIIIa)
Procedure:
- Culture reporter cells to logarithmic growth phase.
- Seed cells in 96-well plates at 50,000 cells/well.
- Treat cells with monomer or aggregate fractions across concentration range (e.g., 0.1-10 µg/mL).
- Incubate plates at 37°C, 5% CO₂ for 20-24 hours.
- Measure GFP fluorescence intensity by flow cytometry or plate reader.
- Calculate fold induction relative to untreated controls and compare dose-response curves between monomeric and aggregated antibodies [71].
Signaling Pathways and Experimental Workflows
Diagram 1: Experimental workflow for assessing aggregation impact on FcγR binding
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagents for Aggregation and FcγR Binding Studies
| Reagent / Solution | Function / Application | Example Specifications |
|---|---|---|
| Therapeutic mAb | Effector-competent model antibody for forced degradation | IgG1, 35-150 mg/mL in histidine buffer, pH 6.0 [71] |
| SEC-HPLC System | Separation and quantification of monomer and aggregate species | TSK-Gel G3000SWxl column, 50 mM NaHâ‚‚POâ‚„, 300 mM NaCl, pH 7.0 mobile phase [71] |
| Surface Plasmon Resonance | Quantitative analysis of FcγR binding affinity and kinetics | Biacore CMS chips, HBS-EP running buffer, FcγR immobilization [71] |
| FcγR Reporter Cells | Functional assessment of receptor activation | Engineered Jurkat cells with NF-κB-GFP reporter [71] |
| Oxidative Stressors | Induction of controlled antibody aggregation | Hydrogen peroxide (1-10 ppm), metal ions (Fe, Cu at 0.5-20 ppm) [71] |
Antibody aggregation presents a complex challenge for therapeutic development, significantly altering FcγR binding profiles in both assay format-dependent and receptor-specific manners. The experimental approaches outlined herein enable systematic evaluation of these effects, from controlled stress induction and aggregate characterization to comprehensive binding and functional assessment. Implementation of these protocols allows researchers to identify critical quality attributes, establish appropriate control strategies, and ultimately ensure the consistent safety and efficacy profile of antibody-based therapeutics throughout their development lifecycle and commercial shelf-life.
Affinity Maturation and Specificity Engineering for Improved Target Engagement
In the competitive landscape of biopharmaceuticals, the development of therapeutic antibodies that exhibit high affinity and exquisite specificity for their targets is paramount for clinical and commercial success. Affinity maturation, the process of enhancing the binding strength between an antibody and its antigen, has emerged as a critical step in antibody engineering pipelines [75]. Concurrently, specificity engineering ensures that this binding is highly selective, minimizing off-target effects and improving therapeutic safety [7]. Together, these processes directly impact target engagement, influencing both the efficacy of the biologic and the required dosing regimen for patients [75] [7]. With clinical trial failures, particularly in late stages, carrying costs that can reach hundreds of millions of dollars, investing in robust affinity and specificity optimization is no longer optional but a necessity for biopharma companies aiming to maintain competitive pipelines [75]. This Application Note provides detailed protocols and strategic frameworks for implementing affinity maturation and specificity engineering, contextualized within the broader thesis of protein engineering for therapeutic antibodies.
Background and Significance
The Molecular Basis of Antibody Affinity
Antibody affinity is quantitatively defined by the equilibrium dissociation constant (KD), which measures the binding strength between an antibody and its target epitope [76]. A lower KD value indicates higher affinity, reflecting tighter and more specific binding. During natural immune responses, B-cells undergo affinity maturation through somatic hypermutation (SHM) and clonal selection in the germinal centers, a process that in vivo can improve antibody affinity by up to 1,000-fold or more from a typical germline KD of ~10-5-10-6 M to mature KD values of ~10-9-10-11 M [76] [77]. In vitro, protein engineering aims to replicate and surpass this natural optimization.
The antigen-binding site of an antibody is formed by six Complementarity-Determining Regions (CDRs) - three each from the heavy (VH) and light (VL) chain variable domains [78]. While traditionally the entire CDR was considered the paratope, structural analyses reveal that only a subset of residues within these loops typically contributes directly to antigen binding [79] [77]. For instance, in anti-NP antibodies, residues at positions 50H, 58H, and 96L directly form hydrogen bonds with the hapten, with the mutation from Lys58H to Arg58H during maturation proving critical for enhancing affinity [77].
The Imperative for Engineering
The business and clinical drivers for affinity maturation are compelling. Higher affinity antibodies can achieve the same therapeutic effect with lower doses, potentially improving patient compliance through reduced administration frequency and minimizing side effects [75]. From a development perspective, candidates with optimized binding are less likely to fail in costly late-stage clinical trials due to insufficient efficacy [75]. Furthermore, as of 2024, approximately 376 antibody therapies have received regulatory approval worldwide, with hundreds more in development, creating intense pressure for product differentiation [80]. In this crowded marketplace, even small advantages in potency or specificity can determine commercial success.
Quantitative Analysis of Affinity Maturation Methods
Table 1: Comparison of Major Affinity Maturation Platforms
| Method | Theoretical Library Size | Key Advantages | Key Limitations | Typical Affinity Gains |
|---|---|---|---|---|
| Phage Display | 1010-1011 clones [6] | Well-established, high diversity, cost-effective [6] | Limited by transformation efficiency, panning artifacts possible | 10-1000x (KD improvements from μM to nM range) [76] |
| Yeast Display | 107-109 clones [75] | Eukaryotic expression, FACS-enabled quantitative screening [76] | Lower library diversity than phage display | 10-1000x (KD improvements from μM to nM range) |
| Ribosome/mRNA Display | 1012-1014 clones [6] | No transformation limit, truly in vitro | Complex biochemistry, requires specialized expertise | Can achieve pM affinity [6] |
| AI/Computational Design | N/A (virtual libraries) | Rapid, targeted, reduces experimental burden [75] [76] | Requires structural data, prediction inaccuracies possible | Varies; can achieve nM-pM affinity when combined with experimental validation |
Table 2: Key Analytical Methods for Affinity Measurement
| Technique | Principle | Throughput | Information Obtained | Sensitivity |
|---|---|---|---|---|
| Surface Plasmon Resonance (SPR) [76] | Label-free detection of binding via refractive index changes | Medium | Kinetic parameters (kon, koff), KD | High (pM-nM KD) |
| Bio-Layer Interferometry (BLI) [76] | Label-free detection via interferometry from biosensor tip | Medium-high | Kinetic parameters (kon, koff), KD | High (pM-nM KD) |
| Isothermal Titration Calorimetry (ITC) [77] | Measures heat changes during binding | Low | Thermodynamic parameters (ΔH, ΔS, KD, stoichiometry) | Medium (nM-μM KD) |
| Enzyme-Linked Immunosorbent Assay (ELISA) [76] | Semi-quantitative colorimetric assay | High | Relative affinity/EC50 | Medium (nM-μM KD) |
Experimental Protocols
Protocol 1: Phage Display-Based Affinity Maturation
Principle: Filamentous phage (M13) display antibody fragments (scFv or Fab) on their surface as fusions to the pIII coat protein, enabling physical linkage between phenotype and genotype [6]. Sequential rounds of biopanning enrich for clones with desired binding characteristics.
Workflow:
Materials:
- Phage display vector (e.g., pHEN series for scFv display)
- E. coli strains: TG1 for phage propagation, HB2151 for soluble expression
- Antigen: 5-100 µg per panning round, purified (>90%)
- Coating buffer: 50 mM NaHCO3, pH 9.6
- Washing buffers: PBS + 0.1% Tween-20, PBS alone
- Elution buffer: 100 mM triethylamine or 0.1 M HCl (for acid elution)
- Neutralization buffer: 1 M Tris-HCl, pH 7.4
Detailed Procedure:
Step 1: Library Construction (3-4 weeks)
- Introduce diversity into parental antibody genes using:
- Error-prone PCR: Use Taq polymerase with unbalanced dNTPs (0.2 mM dATP/dGTP, 1 mM dCTP/dTTP) + 0.5 mM MnCl2 to achieve 1-2 mutations/kb [76].
- CDR-targeted mutagenesis: Design degenerate oligonucleotides to randomize specific CDR residues, prioritizing those identified through structural analysis.
- Clone mutated fragments into phagemid vector via restriction digestion/ligation or Gibson assembly.
- Transform E. coli TG1 by electroporation to achieve library size >109 individual clones. Validate diversity by Sanger sequencing of 20-50 random clones.
Step 2: Panning Cycle (1 week per round)
- Coating: Immobilize 10-100 µg antigen in coating buffer on immunotube or magnetic beads overnight at 4°C. Include no-antigen control for counter-selection.
- Blocking: Block with 2% MPBS (skim milk in PBS) for 2 hours at RT.
- Incubation: Add ~1012 phage particles in 2% MPBS, incubate for 1-2 hours with gentle rotation.
- Washing: Perform 10-20 washes with PBST (PBS + 0.1% Tween-20) followed by 10 washes with PBS. Increase stringency in subsequent rounds by increasing Tween-20 concentration to 0.5%.
- Elution: Incubate with 1 mL triethylamine for 10 minutes with rotation. Neutralize immediately with 0.5 mL Tris-HCl.
- Amplification: Infect log-phase TG1 E. coli with eluted phage. Rescue with helper phage (M13K07) to produce phage for next round.
Step 3: Characterization (2-3 weeks)
- Polyclonal phage ELISA: After rounds 2, 3, and 4, assess enrichment by comparing signal to no-antigen control.
- Monoclonal screening: Pick 96 individual clones for soluble expression. Screen by ELISA and sequence top binders.
- Affinity measurement: Express soluble Fab/scFv of selected clones. Determine KD values using SPR or BLI.
Troubleshooting:
- No enrichment: Reduce washing stringency; verify antigen integrity and coating efficiency.
- Background binding: Incorporate pre-clearing step against bare surface; increase blocking time.
- Limited diversity: Sequence more clones; reduce panning rounds to prevent dominance by early binders.
Protocol 2: Computational Design and Validation
Principle: Using structural models and machine learning to predict affinity-enhancing mutations, reducing experimental screening burden [75] [76].
Workflow:
Materials:
- Structural data: Crystal structure of antibody-antigen complex or high-quality homology models
- Software: Molecular docking (AutoDock Vina, Schrödinger), MD simulation (GROMACS, AMBER), antibody-specific tools (AbRSA, PyIgClassify)
- Hardware: High-performance computing cluster for MD simulations
Detailed Procedure:
Step 1: Model Preparation (1-2 weeks)
- Obtain or generate 3D structure of antibody-antigen complex.
- If using homology modeling, employ tools like AlphaFold2 or RoseTTAFold with antibody-specific constraints.
- Validate model quality using MolProbity; ensure CDR loops are properly modeled.
Step 2: In Silico Mutagenesis (1 week)
- Identify residues for mutation: Prioritize CDR contacts, especially those with high B-factors indicating flexibility.
- Generate single and double mutants in silico using Rosetta or FoldX.
- For each mutant, perform energy minimization to relieve steric clashes.
Step 3: Binding Affinity Prediction (2-3 weeks)
- Run molecular dynamics simulations (50-100 ns) for wild-type and top mutant complexes.
- Calculate binding free energy (ΔGbind) using MM/PBSA or MM/GBSA methods.
- Rank mutants by predicted ΔΔG relative to wild-type.
Step 4: Experimental Validation (3-4 weeks)
- Synthesize genes for top 20-50 predicted mutants.
- Express and purify antibodies in mammalian system (HEK293 or CHO).
- Determine kinetic parameters (kon, koff) and KD using SPR.
- Iterate: Use experimental data to refine computational models.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagent Solutions for Affinity Maturation
| Reagent/Category | Specific Examples | Function/Application | Technical Notes |
|---|---|---|---|
| Display Platforms | M13 phage display [6], yeast surface display [76] | Library creation and screening | Yeast display enables FACS sorting for affinity; phage offers largest library sizes |
| Expression Systems | HEK293, CHO cells [76] | Recombinant antibody production | Mammalian systems ensure proper folding and glycosylation |
| Affinity Measurement Instruments | Biacore (SPR) [76], Octet (BLI) [76] | Quantitative binding kinetics | SPR provides more precise kinetics; BLI offers higher throughput |
| Mutagenesis Kits | Error-prone PCR kits, site-directed mutagenesis kits | Library generation | Commercial kits ensure controlled mutation rates |
| Cell Lines | TG1 E. coli, EBY100 yeast | Display host organisms | TG1 enables efficient phage propagation; EBY100 optimized for surface display |
| Analysis Software | AbYsis [79], PyMol, Rosetta | Structural analysis and design | AbYsis provides antibody-specific numbering and analysis |
Case Study: Structural Insights from Anti-NP Antibodies
Research on anti-(4-hydroxy-3-nitrophenyl)acetyl (NP) antibodies provides exceptional insight into affinity maturation mechanisms [77]. Structural comparisons between germline (N1G9) and affinity-matured (C6, E11) antibodies reveal that:
- The Lys58H to Arg58H mutation in H-CDR2 enhances hydrogen bonding with the NP hapten.
- Introduction of a disulfide bond between Cys96H and Cys100H in E11 stabilizes H-CDR3 conformation, contributing to significantly higher affinity (KD of 5.8 × 108 M-1 vs. C6's 3.3 × 107 M-1).
- Despite 17-24 somatic mutations between germline and matured antibodies, only a handful directly contribute to affinity gains.
Thermodynamic analysis by ITC demonstrates that affinity-matured antibodies often exhibit lower intrinsic stability but achieve remarkable stabilization upon antigen binding, suggesting engineered flexibility enhances binding capacity [77]. This principle should inform engineering strategies—maximizing affinity sometimes requires tolerating moderate destabilization in the unbound state.
Concluding Perspectives
As therapeutic antibody development evolves, affinity maturation and specificity engineering remain cornerstone technologies. The integration of AI and machine learning with high-throughput experimental methods represents the next frontier, enabling more predictive and efficient optimization [75] [76]. Additionally, the growing appreciation for avidity effects and the engineering of multi-specific molecules creates new opportunities to enhance target engagement beyond simple affinity improvements. By implementing the detailed protocols and strategic frameworks outlined in this Application Note, researchers can systematically develop antibody therapeutics with optimized binding characteristics, ultimately contributing to more effective and safer biologic medicines.
High-Throughput Screening and Machine Learning for Developability Prediction
The development of therapeutic antibodies is a complex process where promising candidates frequently fail during late-stage development due to suboptimal biophysical properties. These developability challenges—including aggregation, high viscosity, instability, and immunogenicity—have traditionally been identified only through extensive experimental characterization, resulting in significant resource expenditure and project delays [81]. The integration of high-throughput screening (HTS) methodologies with machine learning (ML) prediction models now enables early assessment and selection of candidates with favorable developability profiles, substantially reducing attrition rates in biotherapeutic development [82].
This paradigm shift is particularly crucial as antibody modalities become increasingly complex, encompassing bispecifics, multispecific formats, and antibody-drug conjugates that present additional developability hurdles [81]. This application note details established protocols and analytical frameworks for implementing HTS and ML strategies to forecast key developability endpoints, providing researchers with practical methodologies to enhance candidate selection in protein engineering pipelines.
High-Throughput Screening Platforms for Antibody Discovery
High-throughput screening technologies enable the rapid evaluation of thousands to millions of antibody variants, generating the extensive datasets necessary for training robust machine learning models.
Display Technologies for Library Screening
Table 1: Comparison of Major Antibody Display Technologies for HTS
| Platform | Library Size | Throughput Capability | Key Advantages | Primary Applications |
|---|---|---|---|---|
| Phage Display | >10^10 [83] | 10^3-10^4 clones/screen [84] | Robust screening; compatibility with automation and NGS | Hit generation and affinity maturation |
| Yeast Display | 10^7-10^9 [83] | 10^8 cells screened [85] | Eukaryotic secretion machinery; proper folding and PTMs | Affinity maturation and stability engineering |
| Mammalian Cell Display | 10^7-10^9 | High-throughput FACS [84] | Full IgG display; native PTMs | Membrane protein targeting and functional screening |
| B Cell Sorting | N/A (natural repertoire) | 300-5,000 events/second [85] | Native heavy-light chain pairing; no reformatting required | Recovery of natural immune responses |
Protocol: Yeast Surface Display for Antibody Screening
Principle: The yeast display system expresses antibody fragments (e.g., scFv, Fab) fused to agglutinin proteins on the yeast cell surface, enabling quantitative screening using fluorescence-activated cell sorting (FACS) or microfluidics [85] [84].
Materials:
- Induced yeast library expressing antibody variants
- Fluorescently-labeled antigen (e.g., biotinylated antigen + streptavidin-PE)
- FACS sorter or microfluidic screening system
- Selection media (SD/-Trp, SD/-Trp/-Ura)
Procedure:
- Library Induction: Induce yeast library expression in SG/-Trp media at 20-30°C for 24-48 hours.
- Labeling: Incubate 10^7-10^8 yeast cells with fluorescently-labeled antigen (0.1-100 nM range) for 30-60 minutes on ice.
- Washing: Pellet cells and wash twice with PBS + 0.1% BSA to remove unbound antigen.
- Sorting: Sort top 0.1-5% of fluorescent population using FACS or microfluidics.
- Recovery: Grow sorted populations in selective media for 24-48 hours.
- Iteration: Repeat steps 1-5 for 2-4 rounds with increasing stringency (decreased antigen concentration).
- Clone Isolation: Plate final sorted population for single colony isolation and sequence analysis.
Automation Enhancement: Implement robotic liquid handling systems to automate labeling and washing steps, increasing throughput to 10^8 variants per screening round [86].
Microfluidics and Compartmentalization Strategies
Advanced compartmentalization technologies enable ultra-high-throughput screening by miniaturizing reaction volumes to the picoliter scale, dramatically increasing screening efficiency while reducing reagent consumption [85].
Table 2: Compartmentalized Screening Platforms for Antibody Engineering
| Compartment Type | Volume Range | Throughput (events/sec) | Example Application | Result |
|---|---|---|---|---|
| Water-in-Oil Droplets | Picoliters [85] | 2,000-20,000 [85] | Enzyme variant screening | 160% increase in inhibitor resistance [85] |
| Gel-Shell Beads (GSBs) | Microliters [85] | ~2,800 [85] | Phosphotriesterase engineering | 19-fold increase in kcat/KM [85] |
| FurShell | Microliters [85] | 5,000 [85] | Phytase thermostability | 97 U·mgâ»Â¹ higher specific activity [85] |
| CHESS | Microliters [85] | 8,000 [85] | GPCR thermostability | ~26.8°C increase in Tm [85] |
Protocol: Drop-Based Microfluidics for Antibody Screening
Principle: Water-in-oil emulsion droplets function as picoliter-volume compartments for individual antibody expression and functional assessment, enabling analysis of >10^6 variants per day [85].
Materials:
- Microfluidic droplet generation system
- In vitro transcription/translation system
- Fluorogenic substrate or detecting reagent
- Surfactants and oil phase
- Droplet sorting instrumentation
Procedure:
- Library Compartmentalization: Combine antibody DNA library with in vitro expression system and substrate, then inject into droplet generator to create monodisperse droplets.
- Incubation: Incubate emulsions at 30°C for 2-4 hours to enable protein expression.
- Detection: Monitor fluorescence development within droplets using inline detection.
- Sorting: Activate droplet sorter to isolate hits based on fluorescence intensity (≥502-fold enrichment achievable) [85].
- Recovery: Break emulsion and recover DNA from sorted droplets for sequencing and validation.
Machine Learning for Developability Prediction
Machine learning leverages high-throughput screening data to build predictive models for key developability properties, enabling in silico candidate prioritization before expensive experimental characterization.
Feature Engineering and Model Training
Critical Developability Endpoints:
- Hydrophobicity: Predictive of aggregation and non-specific binding
- Poly-specificity: Propensity for off-target interactions
- Stability: Thermal and conformational stability
- Immunogenicity: Potential to elicit anti-drug antibodies [82]
Protocol: Developing ML Models for Developability Prediction
Principle: Machine learning algorithms identify patterns between computationally-derived antibody sequence features and experimentally-measured developability parameters [82].
Materials:
- Curated antibody sequence-structure dataset (≥100 variants)
- Molecular modeling software (e.g., for calculating structural descriptors)
- Machine learning environment (Python/R with scikit-learn, TensorFlow)
- High-performance computing resources
Procedure:
- Dataset Curation: Compile dataset with paired antibody sequences and experimental developability measurements.
- Feature Extraction: Calculate diverse feature sets including:
- Sequence-based features (amino acid composition, physico-chemical properties)
- Structure-based features (solvent accessible surface area, electrostatic potentials)
- Dynamics-based features (molecular flexibility, aggregation-prone regions)
- Feature Selection: Apply automated feature selection algorithms (e.g., recursive feature elimination) to identify most predictive descriptors.
- Model Training: Implement multiple algorithms (random forest, gradient boosting, neural networks) using cross-validation.
- Model Validation: Evaluate performance on held-out test set using metrics (R², RMSE, AUC).
- Deployment: Integrate best-performing model into discovery workflow for prospective prediction.
Key Descriptors for Developability Prediction
Table 3: Important in silico Descriptors for Developability Prediction
| Descriptor Category | Specific Features | Correlation with Developability |
|---|---|---|
| Sequence-Based | Hydrophobicity indices, charge distribution, cysteine content | Hydrophobicity correlates with aggregation risk; net charge affects viscosity [82] |
| Structural | Solvent accessible surface area, secondary structure propensity | Structural stability measurements predict expression yield [82] |
| Dynamic | Molecular flexibility, B-factors, hydrogen bonding patterns | Flexibility in CDR regions can indicate conformational instability |
| Surface Properties | Electrostatic potentials, patch analysis | Surface hydrophobicity patches predict polyspecificity [82] |
Integrated Workflow: From Screening to Prediction
Successful implementation requires seamless integration of experimental screening with computational prediction in an iterative feedback loop.
Figure 1: Integrated workflow combining high-throughput experimental screening with machine learning prediction for antibody developability assessment. The feedback loop continuously improves model performance as additional experimental data is generated.
Essential Research Reagent Solutions
Table 4: Key Research Reagents for HTS and Developability Assessment
| Reagent/Technology | Supplier Examples | Application in Workflow |
|---|---|---|
| High-Throughput Gene Synthesis | Twist Bioscience, Thermo Fisher [87] | Rapid generation of variant libraries for screening |
| HTP Antibody Expression Service | Thermo Fisher [87] | Parallel production of hundreds of antibody candidates |
| Automated Screening Platforms | Opentrons, Genedata [86] | Laboratory automation for increased throughput |
| Display System Vectors | Neochromosome [88] | Yeast display systems with switchable display/secretion |
| Biosensor Tools | Sartorius (Octet) [84] | High-throughput kinetic characterization |
| NGS Library Prep Kits | Illumina | Deep sequencing of antibody libraries |
The strategic integration of high-throughput screening technologies with machine learning-based prediction represents a transformative approach to addressing developability challenges in therapeutic antibody development. The protocols and methodologies detailed in this application note provide researchers with practical frameworks for implementation, potentially reducing late-stage attrition and accelerating the development of next-generation biotherapeutics. As these technologies continue to evolve, particularly with advances in microfluidics and artificial intelligence, the ability to forecast and engineer favorable developability characteristics will become increasingly sophisticated and integral to successful antibody discovery campaigns.
From Bench to Bedside: Clinical Translation and Commercial Landscape
Therapeutic antibodies have revolutionized modern medicine, offering targeted treatments for a wide spectrum of diseases, including cancer, autoimmune disorders, and migraines. As of 2025, over 200 antibody-based therapeutics have been marketed globally, with a robust pipeline of nearly 1,400 investigational candidates [89]. The success of these biologics is deeply rooted in advanced protein engineering, which has transformed native antibody structures into highly effective drugs with enhanced properties. This application note details the engineering mechanisms behind key clinically approved antibodies, providing structured data and methodologies to inform research and development efforts.
Clinically Approved Engineered Antibodies
Engineering strategies have been critical in developing antibodies with improved efficacy, stability, and pharmacokinetics. The following section highlights seminal successes in antibody engineering.
Key Approved Engineered Antibodies
Table 1: Clinically Approved Engineered Antibodies and Their Engineering Mechanisms
| Antibody (Brand Name) | Target | Key Indications | Engineering Mechanism | Effect of Engineering |
|---|---|---|---|---|
| Adalimumab (Humira) [90] | TNF-α | Rheumatoid Arthritis, Psoriasis, IBD | Human monoclonal antibody; pioneered the use of fully human antibodies for chronic inflammation. | High specificity for TNF-α; reduced immunogenicity compared to earlier chimeric or humanized antibodies. |
| Ravulizumab (Ultomiris) [7] | Complement C5 | Paroxysmal Nocturnal Hemoglobinuria, aHUS | Fc engineering with M428L/N434S (LS) mutations in the Fc region. | Increased half-life (~4x longer) by enhancing pH-dependent binding to FcRn for more efficient recycling; allows for longer dosing intervals. |
| Zanidatamab (Ziihera) [89] | HER2 (bispecific) | HER2-positive cancers | Bispecific antibody engineered to bind two distinct epitopes on the HER2 receptor. | Enhanced tumor growth inhibition by simultaneous dual-epitope engagement and reduced potential for drug resistance. |
| Ivosidenib [89] | Not Specified | Not Specified | Antibody-Drug Conjugate (ADC) combining a target-specific antibody with a cytotoxic drug. | Enables targeted delivery of a potent chemotherapeutic directly to cancer cells, maximizing efficacy and minimizing systemic toxicity. |
| Insulin Glargine [7] | Insulin Receptor | Diabetes | Amino acid modifications (Asn21→Gly in A-chain; C-terminal Arg addition on B-chain). | Altered pI leads to precipitation at injection site, providing a slow-release depot effect and prolonged duration of action (up to 24 hours). |
| Teplizumab (Tzield) [90] | CD3 | Delay of Type 1 Diabetes | Engineered to modulate T-cell function. | First disease-modifying therapy for Type 1 diabetes; delays clinical onset by targeting autoimmune T-cells. |
Analysis of Engineering Trends
The late-stage clinical pipeline continues to emphasize innovative formats. As of late 2024, over 20 bispecific antibodies and numerous Antibody-Drug Conjugates (ADCs) were in advanced development or regulatory review [89]. Promising late-stage candidates include denecimig and sonelokimab, demonstrating the field's move towards multispecificity and novel mechanisms of action [89]. However, developability remains a key challenge. A 2024 comparative study demonstrated that while engineered formats like bispecifics and scFvs offer significant therapeutic potential, they often face greater instability, aggregation, and fragmentation issues compared to the natural full-length IgG format [91]. This underscores the critical need for robust engineering and formulation strategies to translate complex designs into successful medicines.
Experimental Protocols for Antibody Engineering and Analysis
This section provides detailed methodologies for key experiments in antibody development, from initial design to stability assessment.
Protocol: In Silico Affinity Maturation
Purpose: To enhance antibody-antigen binding affinity through computational design. Applications: Improving potency of lead therapeutic antibody candidates.
Procedure:
- Structure Preparation: Obtain a high-resolution crystal structure of the antibody-antigen (Fab-Ag) complex. If unavailable, generate a high-quality homology model [92].
- In Silico Mutagenesis: Systematically mutate residues within the Complementarity-Determining Regions (CDRs) to all other 19 natural amino acids.
- Energy Evaluation:
- Perform an initial rigid-body docking and side-chain rotamer search to identify low-energy conformations.
- Re-evaluate top candidates using more accurate, computationally expensive methods like Molecular Dynamics (MD) simulations or Poisson-Boltzmann (PB) continuum electrostatics to calculate binding free energy [92].
- Candidate Selection: Select mutant variants exhibiting the most favorable (lowest) computed interaction energy with the antigen.
- Experimental Validation: Express the top in silico-designed antibody variants and measure binding affinity using Surface Plasmon Resonance (SPR). A successful campaign can achieve a 10-fold or greater increase in affinity [92].
Protocol: Developability Assessment via Stability and Aggregation Propensity
Purpose: To evaluate the biophysical stability and aggregation propensity of engineered antibody formats, a critical step for candidate selection [91]. Applications: Profiling and ranking lead candidates, especially non-standard formats like scFvs and bispecifics.
Procedure:
- Sample Preparation: Purify the antibody construct using affinity chromatography (e.g., Protein A) followed by size-exclusion chromatography (SEC) to isolate the monomeric fraction.
- Purity Analysis (SE-HPLC):
- Inject the purified sample onto an SEC column.
- Analysis: Calculate the percentage of monomeric protein. Purity above 95% is considered low risk, while values below 90% indicate high developmental risk [91].
- Conformational Stability (Differential Scanning Calorimetry - DSC):
- Ramp the temperature of the antibody solution (e.g., 0.5-1.0°C/min) while measuring heat flow.
- Analysis: Determine the melting temperature (Tm) of the domains. A Tm > 60°C is considered low risk; a Tm < 55°C indicates instability [91].
- Colloidal Stability (Aggregation Temperature - Tagg):
- Use a method like static light scattering (SLS) while ramping the sample temperature.
- Analysis: Identify the temperature at which aggregation onset (Tagg) occurs. A Tagg > 60°C is desirable [91].
- Stress Testing (Agitation & Interfacial Stress):
- Agitate the protein sample vigorously or expose it to air-water interfaces.
- Analysis: Quantify monomer loss via SEC-HPLC after stress. Formats with a higher propensity for aggregation and fragmentation will show greater monomer loss [91].
Research Reagent Solutions
Table 2: Essential Reagents and Materials for Antibody Development
| Reagent / Material | Function / Application | Brief Description |
|---|---|---|
| Surface Plasmon Resonance (SPR) | Binding affinity (KD) and kinetics (kon, koff) measurement | A biosensor technique for label-free, real-time analysis of biomolecular interactions (e.g., antibody-antigen binding) [91]. |
| Size-Exclusion HPLC (SE-HPLC) | Purity and aggregation analysis | Chromatographic method to separate and quantify monomeric antibodies from high-molecular-weight aggregates and fragments [91]. |
| Differential Scanning Calorimetry (DSC) | Conformational thermal stability | Measures the thermal stability of protein domains by detecting heat absorption during unfolding, providing melting temperatures (Tm) [91]. |
| Knob-into-Hole Fc Technology | Bispecific antibody assembly | An engineered Fc heterodimerization strategy using complementary mutations in the CH3 domains to ensure correct heavy chain pairing [91]. |
| Stabilizing Disulfide Bond (VH44-VL100) | scFv and fragment stability | Introduction of an engineered disulfide bond between the variable heavy and light chains to improve scFv stability and prevent dissociation [91]. |
| FcRn Binding Assay | Pharmacokinetic half-life assessment | In vitro assay (e.g., SPR) to measure antibody binding to the Neonatal Fc Receptor at endosomal pH (~6.0), predicting in vivo half-life [91]. |
Visualizing Antibody Engineering Workflows
Antibody Structure and Engineering Sites
Antibody Engineering and Developability Workflow
The development of therapeutic antibodies has been revolutionized by advances in protein engineering, enabling the creation of highly specific, potent, and safe biologics for a wide range of diseases. Since the introduction of hybridoma technology in 1975, the field has witnessed a succession of innovations including chimeric and humanized antibody engineering, phage display, transgenic mouse platforms, and high-throughput single B cell isolation [8]. These technological developments have enhanced the specificity, potency, and safety of monoclonal antibodies (mAbs), resulting in 144 FDA-approved antibody drugs on the market and 1,516 worldwide candidates in clinical development as of August 2025 [8]. Engineering breakthroughs have led to new modalities of antibody-based therapeutics, such as antibody-drug conjugates (ADCs), bispecific antibodies (bsAbs), and chimeric antigen receptor T (CAR-T) cell therapies, each with therapeutic utility across multiple disease domains [8]. This application note provides a comparative analysis of major antibody engineering platforms, detailing their strengths, limitations, and experimental protocols to guide researchers in selecting appropriate technologies for therapeutic antibody development.
Platform Comparisons: Quantitative Analysis
The antibody discovery services market, valued at USD 1.90 billion in 2025, is projected to advance at a CAGR of 13.3% from 2025 to 2030, reaching USD 3.54 billion [93]. This growth is propelled by technological advancements, increasing demand for biologics, and the development of new antibody formats. Below are structured comparisons of key engineering platforms.
Table 1: Comparison of Major Antibody Discovery Platforms
| Platform | Key Features | Therapeutic Success | Development Timeline | Relative Cost |
|---|---|---|---|---|
| Hybridoma | - Murine-derived antibodies- Immunization-based- High immunogenicity risk | 34% of approved mAbs [8] | 3-6 months | $$ |
| Phage Display | - Fully human antibodies- In vitro selection- Bypasses immunization | 16 FDA-approved drugs [8] | 2-4 months | $$$ |
| Transgenic Mice | - Fully human antibodies- In vivo maturation- Preserves natural pairing | 30 fully human antibodies & 3 bsAbs approved [8] | 6-9 months | $$$$ |
| Single B Cell Screening | - Fully human antibodies- High-throughput- Preserves native pairs | Emerging (SARS-CoV-2, HIV) [8] | 1-3 months | $$$$ |
Table 2: Analysis of Antibody Engineering Strategies for Optimizing Therapeutic Properties
| Engineering Strategy | Mechanism of Action | Impact on Developability | Clinical Example |
|---|---|---|---|
| Affinity Maturation | - Site-directed mutagenesis- CDR optimization- Library screening | Increases binding affinity (K_D); can affect specificity | Ranibizumab (Lucentis) for VEGF-A [94] |
| Fc Engineering | - Mutations in Fc region- Altered FcγR binding- Modulated FcRn interaction | Enhances half-life, effector functions; reduces immunogenicity | Ravulizumab (Ultomiris) with extended half-life [7] |
| Humanization | - CDR grafting- Framework optimization- Resurfacing | Reduces HAMA response; maintains binding affinity | Trastuzumab (Herceptin) for HER2+ breast cancer [8] |
| Bispecific Formatting | - Dual-targeting scFv- Cross-arm coordination- Asymmetric design | Enables novel mechanisms; complex manufacturability | Emicizumab (Hemlibra) for hemophilia A [8] |
Established and Emerging Engineering Strategies
Site-Specific Mutagenesis for Stability and Function
Site-specific mutagenesis represents a fundamental protein engineering approach for enhancing therapeutic properties of antibodies. A classic application involves developing insulin variants with different kinetics of action [7]. For instance:
- Insulin glargine: Substitution of asparagine by glycine at amino acid 21 of the α chain and addition of 2 arginines to the β chain increases the isoelectric point (pI), resulting in precipitation upon injection and prolonged duration of action up to 24 hours [7].
- Insulin glulisine: Exchange of β chain asparagine (position 3) and lysine (position 29) with lysine and glutamic acid, respectively, decreases pI from 5.5 to 5.1, promoting increased solubility with less propensity for hexamer formation and fast-acting effect [7].
Targeted mutations also significantly enhance antibody stability and pharmacokinetics. The spatial aggregation propensity (SAP) method, a molecular dynamics-based simulation technique, identifies key regions in proteins that drive aggregation, enabling specific mutations that enhance stability compared to the native protein [7]. Fc engineering with mutations such as M428L/N434S (LS variant) and M252Y/S254T/T256E (YTE variant) modulate binding to the neonatal Fc receptor (FcRn), extending serum half-life by promoting antibody recycling rather than lysosomal degradation [7].
Phage Display Technology for Antibody Discovery
Phage display technology has revolutionized antibody discovery by enabling fully human antibody generation without immunization. Since its development in 1985 by George P. Smith, phage display has evolved through three generations of library designs [94]. The technology works by displaying antibody fragments (scFv, Fab, VHH) on the surface of bacteriophages, with the genetic information encoded within the phage particle, creating a physical link between phenotype and genotype [94].
Experimental Protocol: Phage Display Library Panning
Reagents Required:
- Phage display library (e.g., scFv or Fab library)
- Target antigen (purified protein, peptides, or cell-bound)
- Coating buffers (e.g., carbonate-bicarbonate buffer, pH 9.6)
- Blocking buffer (e.g., 2-5% BSA or milk in PBS)
- Washing buffers (PBS with 0.1% Tween-20)
- Elution buffer (e.g., 0.1 M glycine-HCl, pH 2.2 with 1 mg/mL BSA)
- Neutralization buffer (e.g., 1 M Tris-HCl, pH 9.0)
- E. coli host strains (e.g., TG1 or XL1-Blue)
- Growth media (2x YT with appropriate antibiotics)
Procedure:
- Antigen Coating: Immobilize 10-100 μg of purified target antigen in coating buffer overnight at 4°C on immunotubes or plates. Alternatively, use biotinylated antigen in solution with streptavidin-coated magnetic beads.
- Blocking: Block coated surfaces with 2-5% BSA or milk in PBS for 1-2 hours at room temperature to reduce non-specific binding.
- Phage Binding: Incubate phage library (10¹¹-10¹³ CFU) in blocking buffer with the antigen for 1-2 hours at room temperature with gentle agitation.
- Washing: Remove unbound phages through extensive washing (10-20 washes) with PBS containing 0.1% Tween-20, followed by PBS alone.
- Elution: Bound phages are eluted using 0.1 M glycine-HCl (pH 2.2) for 10 minutes, then neutralized with 1 M Tris-HCl (pH 9.0). Alternatively, use competitive elution with target antigen or trypsin digestion.
- Amplification: Infect log-phase E. coli (e.g., TG1) with eluted phages, culture in 2x YT media with appropriate antibiotics, and rescue with helper phages (e.g., M13K07) to produce phage particles for subsequent rounds of panning.
- Selection Monitoring: Typically perform 3-4 rounds of panning, monitoring enrichment through phage ELISA or output titer calculations.
This protocol has enabled the development of 17 FDA-approved therapeutic antibodies, including adalimumab (Humira), the first fully human antibody developed via phage display [94] [8].
Transgenic Mouse Platforms
Transgenic mouse platforms, introduced in 1994 with the HuMab Mouse and XenoMouse, incorporate human immunoglobulin genes to enable generation of fully human antibodies following immunization [8]. These platforms preserve the natural in vivo affinity maturation process while producing antibodies with reduced immunogenicity.
Experimental Protocol: Immunization and Hybridoma Generation from Transgenic Mice
Reagents Required:
- Transgenic mice (e.g., HuMab, XenoMouse, VelocImmune)
- Antigen of interest (≥90% purity recommended)
- Adjuvants (e.g., Freund's, Alum, or TLR agonists)
- Myeloma cells (e.g., SP2/0 or P3X63Ag8.653)
- Polyethylene glycol (PEG) solution for fusion
- HAT selection medium
- ELISA plates and detection reagents
Procedure:
- Immunization: Immunize 6-8 week old transgenic mice with 10-50 μg antigen emulsified in appropriate adjuvant via subcutaneous, intraperitoneal, or footpad routes. Boost every 2-3 weeks for 2-4 cycles.
- Serum Titer Monitoring: Collect serum 7-10 days post-immunization and evaluate antigen-specific antibody responses by ELISA.
- Fusion Preparation: Administer final boost 3-4 days before fusion. Harvest splenocytes and mix with myeloma cells at 4:1 ratio.
- Cell Fusion: Centrifuge cell mixture and fuse with 50% PEG solution over 1 minute. Gradually dilute PEG and wash cells.
- Hybridoma Selection: Plate fused cells in HAT selection medium to eliminate unfused myeloma cells. Feed cells with fresh HAT medium every 2-3 days.
- Screening: Screen supernatants for antigen-specific antibodies by ELISA after 10-14 days. Expand positive wells and perform subclass analysis.
- Cloning: Perform limiting dilution cloning to ensure monoclonality. Re-test and expand stable clones for antibody production.
This platform has yielded 30 fully human antibodies with FDA approval, including panitumumab (Vectibix), the first transgenic mouse-derived human antibody drug approved in 2006 for cancer treatment [8].
Single B Cell Screening Technologies
Single B cell antibody screening platforms represent a powerful method to generate fully human monoclonal antibodies, particularly for isolating neutralizing antibodies against infectious diseases [8]. These technologies preserve the natural heavy and light chain pairing while enabling high-throughput screening.
Experimental Protocol: Single B Cell Sorting and Antibody Cloning
Reagents Required:
- Fluorescently-labeled antigens or probes
- Cell sorting buffers (PBS with 1-5% FBS)
- Cell culture media (RPMI-1640 or DMEM)
- Lysis buffer for RNA extraction
- Reverse transcription and PCR reagents
- Expression vectors (e.g., IgG1 backbone)
- Transfection reagents (e.g., PEI, Lipofectamine)
Procedure:
- B Cell Staining: Stain peripheral blood mononuclear cells (PBMCs) or splenocytes with fluorescently-labeled antigen and B cell markers (e.g., CD19, CD20, CD27).
- Single-Cell Sorting: Sort single antigen-specific B cells into 96-well or 384-well plates containing lysis buffer using FACS.
- Reverse Transcription: Perform reverse transcription directly in lysis buffer to generate cDNA.
- Amplification: Amplify antibody variable heavy (VH) and light (VL) chain genes using nested PCR with family-specific primers.
- Cloning: Clone amplified VH and VL fragments into antibody expression vectors containing constant regions.
- Recombinant Expression: Transiently transfect HEK293 or ExpiCHO cells with paired heavy and light chain vectors.
- Screening: Screen supernatants for antigen binding by ELISA or flow cytometry. Expand positive hits for characterization.
Advanced technologies such as droplet-based microfluidics and optofluidics allow high-throughput pairing of VH and VL transcripts from individual B cells, followed by next-generation sequencing for antibody reconstruction, greatly accelerating antibody discovery [8].
Artificial Intelligence in Antibody Engineering
The integration of artificial intelligence (AI) and machine learning (ML) is transforming antibody discovery by enabling faster timelines, reducing manual processes, and improving candidate quality [93] [8]. AI-driven platforms combine machine learning, robotics, and predictive modeling to accelerate lead generation and optimize antibody design.
Recent breakthroughs in AI, particularly structure-prediction tools like AlphaFold-Multimer and AlphaFold 3, have significantly advanced our ability to model antibody-antigen complexes with atomic-level accuracy [8]. In parallel, generative models such as RoseTTAFold and RFdiffusion have enabled de novo design of antibody scaffolds and binding interfaces [8]. These innovations offer powerful new strategies for accelerating antibody development in oncology, infectious disease, and beyond.
Experimental Protocol: AI-Guided Antibody Optimization
Reagents Required:
- Initial antibody sequence and structural data
- High-throughput binding data (e.g., SPR, BLI)
- Computational resources (GPU clusters)
- ML software platforms (e.g., TensorFlow, PyTorch)
- Expression and purification systems for validation
Procedure:
- Data Curation: Collect existing antibody sequences, structural data, and binding affinity measurements to create training datasets.
- Feature Engineering: Extract relevant features including CDR loop lengths, structural parameters, and physicochemical properties.
- Model Training: Train machine learning models (e.g., random forest, neural networks) on curated datasets to predict binding affinity and developability properties.
- In Silico Mutagenesis: Generate virtual antibody variants and predict their properties using trained models.
- Library Design: Select top predicted variants for synthesis and testing.
- Experimental Validation: Express and purify selected variants, then characterize their binding affinity, specificity, and stability.
- Model Refinement: Incorporate experimental results to refine predictive models in an iterative feedback loop.
Companies such as LabGenius apply machine learning with robotics to design and test antibodies with minimal human input, enabling rapid discovery cycles [93]. Absci Corporation, supported by a major collaboration with Merck, leverages predictive modeling to generate and optimize antibodies [93].
Research Reagent Solutions
Table 3: Essential Research Reagents for Antibody Engineering Platforms
| Reagent Category | Specific Examples | Function in Workflow | Technical Notes |
|---|---|---|---|
| Display Libraries | - scFv phage libraries- Fab phage libraries- VHH libraries | Source of antibody diversity for in vitro selection | Library quality critical; diversity of 10¹â°-10¹² CFU recommended [94] |
| Cell Lines | - E. coli TG1/XLI-Blue- HEK293/ExpiCHO- Myeloma cells (SP2/0) | Antibody expression and production | Different hosts affect glycosylation patterns and yield [8] |
| Selection Reagents | - Biotinylated antigens- Streptavidin magnetic beads- Coating buffers | Enrichment of antigen-specific binders | Solution-phase selection reduces conformational bias [94] |
| Detection Systems | - Anti-Fab/FC conjugates- Protein A/G reagents- ELISA substrates | Identification and characterization of binders | High-sensitivity detection enables rare clone identification [95] |
| Cloning Systems | - Restriction enzymes |
Antibody gene manipulation and expression | Modular vector systems enable rapid reformatting [8] |
The comparative analysis of antibody engineering platforms reveals a dynamic landscape where established technologies like hybridoma and phage display continue to evolve alongside emerging approaches such as single B cell screening and AI-driven design. Each platform offers distinct advantages: phage display provides unparalleled control over selection conditions and epitope targeting [94], transgenic mouse platforms preserve natural antibody maturation processes [8], and single B cell technologies maintain native heavy-light chain pairing while enabling rapid discovery [8]. The integration of artificial intelligence and machine learning is further transforming antibody engineering by enabling predictive design and reducing development timelines [93] [8]. As the field advances, the strategic selection and integration of these platforms will be essential for developing next-generation antibody therapeutics with enhanced efficacy, reduced immunogenicity, and improved manufacturability. Researchers should consider their specific target antigen, desired antibody properties, and available resources when selecting the most appropriate engineering platform for their therapeutic development programs.
Pharmacokinetic Profiling and Preclinical Prediction for Fc-Engineered mAbs
Within the broader thesis of protein engineering for therapeutic antibodies, the pharmacokinetic (PK) profiling of Fc-engineered monoclonal antibodies (mAbs) represents a critical step in translating innovative designs into clinically viable therapeutics. Fc engineering, which modifies the fragment crystallizable (Fc) region of an antibody, aims to enhance therapeutic properties, notably by modulating binding to the neonatal Fc receptor (FcRn) to prolong serum half-life and improve exposure [96] [97]. For researchers and drug development professionals, establishing robust and predictive preclinical protocols is paramount to accurately forecasting human PK, thereby de-risking development and accelerating the path to the clinic. This document provides detailed application notes and protocols for the PK profiling and preclinical prediction of human PK for Fc-engineered mAbs using advanced in vivo models and allometric scaling techniques.
Key Quantitative Data and Comparisons
The following tables consolidate critical quantitative findings from a recent study on predicting human PK for Fc-engineered mAbs using human FcRn transgenic mice (Tg32) [96].
Table 1: Key Pharmacokinetic (PK) Parameters and Their Definitions
| PK Parameter | Symbol | Definition |
|---|---|---|
| Clearance | CL | Volume of plasma cleared of the drug per unit time |
| Inter-compartmental Clearance | Q | Distribution clearance between central and peripheral compartments |
| Volume of Distribution (Central) | Vc | Theoretical volume of the central compartment (plasma) |
| Volume of Distribution (Peripheral) | Vp | Theoretical volume of the peripheral compartment (tissues) |
| Half-life | t~1/2~ | Time required for the plasma concentration to reduce by 50% |
Table 2: Optimal Allometric Scaling Exponents for Fc-Engineered mAbs from Tg32 Mice to Humans
| PK Parameter | Allometric Exponent |
|---|---|
| Clearance (CL) | 0.73 |
| Inter-compartmental Clearance (Q) | 0.60 |
| Volume of Distribution - Central (Vc) | 0.95 |
| Volume of Distribution - Peripheral (Vp) | 0.87 |
Table 3: Experimental Conditions in Tg32 Mouse Study and Correlation with Clinical Outcomes
| Experimental Variable | Observation in Tg32 Model | Clinical Correlation |
|---|---|---|
| IVIG Co-administration | Normal mAbs showed higher CL with IVIG; Fc-engineered mAbs showed comparable CL with/without IVIG. | The larger CL difference mirrored clinical study results, validating the model. |
| CL Correlation | A significant positive correlation was observed for Fc-engineered mAbs in both absence and presence of IVIG. | Supports Tg32 mice as a predictive tool for human CL. |
| Overall Prediction Accuracy | The established methodology using optimized exponents accurately predicted human plasma concentration-time profiles. | Achieved prediction accuracy comparable to cynomolgus monkeys. |
Experimental Protocols
Protocol: Pharmacokinetic Study in Human FcRn Transgenic (Tg32) Mice
This protocol describes the procedure for evaluating the pharmacokinetics of Fc-engineered mAbs in the Tg32 mouse model, a critical step for human PK prediction [96].
1. Materials and Reagents
- Test Articles: Fc-engineered mAbs and normal (unengineered) mAbs as controls.
- Animals: Human FcRn transgenic mice (e.g., Tg32 strain).
- Reagents: Intravenous immunoglobulin (IVIG), such as at a dose of 1000 mg/kg.
- Equipment: Sterile syringes and needles, microcentrifuge tubes, anesthesia equipment (if needed), analytical equipment for mAb quantification (e.g., ELISA, MSD).
2. Experimental Design and Dosing
- Formulate mAbs in an appropriate buffer for in vivo administration (e.g., PBS).
- Randomly assign Tg32 mice into experimental groups. Key groups include:
- Fc-engineered mAb, without IVIG.
- Fc-engineered mAb, with IVIG.
- Normal mAb, without IVIG (control).
- Normal mAb, with IVIG (control).
- Administer all mAbs via a single intravenous (IV) bolus injection at a dose of 10 mg/kg.
- For groups receiving IVIG, co-administer IVIG at 1000 mg/kg intravenously.
3. Sample Collection
- Collect blood samples (e.g., via retro-orbital bleeding or tail vein) at predetermined time points post-dose. A typical profile may include time points such as: 5 minutes (post-dose), 4, 8, 24, 48, 72, 96, 120, 168, 240, and 336 hours.
- Centrifuge blood samples to isolate plasma and store at -80°C until analysis.
4. Bioanalytical Analysis
- Quantify the plasma concentration of the administered mAb using a validated method, such as a target-capture or anti-idiotypic ELISA.
- Generate standard curves using the reference standard of the mAb in naïve mouse plasma.
5. PK Data Analysis
- Perform non-compartmental analysis (NCA) on the plasma concentration-time data for each animal to estimate primary PK parameters (e.g., AUC, CL, V~ss~, t~1/2~).
- Alternatively, use the data to develop a population PK model to estimate parameters like CL, Q, Vc, and Vp.
Protocol: Allometric Scaling for Human PK Prediction
This protocol outlines the steps for scaling PK parameters from Tg32 mice to humans to predict human PK profiles [96].
1. Parameter Estimation in Mice
- Using the plasma concentration-time data from the Tg32 mouse study, estimate the fundamental PK parameters (CL, Q, Vc, Vp) via a suitable compartmental modeling approach.
2. Application of Allometric Scaling
- Apply species scaling using the following allometric equation with fixed exponents:
Human Parameter = Mouse Parameter × (Human Body Weight / Mouse Body Weight)^Exponent^
- Use the optimized exponents specific for Fc-engineered mAbs as listed in Table 2. Assume a standard mouse body weight of 0.025 kg and a standard human body weight of 70 kg.
3. Prediction of Human Plasma Profile
- Incorporate the scaled human PK parameters (CL~human~, Q~human~, Vc~human~, Vp~human~) into a two-compartment PK model.
- Simulate the plasma concentration-time profile following an intravenous dose in humans.
Visualizing Workflows and Mechanisms
FcRn Recycling and PK Prediction Workflow
Experimental and Prediction Logic
The Scientist's Toolkit: Research Reagent Solutions
Table 4: Essential Research Reagents and Materials for Fc-mAb PK Studies
| Item | Function / Application in Protocol |
|---|---|
| Human FcRn Transgenic Mice (Tg32) | The critical in vivo model expressing the human FcRn receptor, enabling predictive assessment of human PK for mAbs with modified Fc regions [96]. |
| Intravenous Immunoglobulin (IVIG) | Used to saturate FcRn receptors in control experiments; it helps differentiate the PK of Fc-engineered mAbs (which resist displacement) from normal mAbs [96]. |
| Fc-engineered mAbs | Test articles with specific mutations (e.g., YTE, LS) in the Fc region designed for enhanced affinity to human FcRn, leading to improved half-life [96] [97]. |
| Anti-idiotypic Capture Assay | A highly specific bioanalytical method (e.g., ELISA) used to accurately quantify the plasma concentration of the administered therapeutic mAb amidst high levels of endogenous IgG [96]. |
| Allometric Scaling Exponents | Empirically derived, fixed exponents for key PK parameters (CL, Q, Vc, Vp) that optimize the accuracy of human PK predictions from Tg32 mouse data for Fc-engineered mAbs [96]. |
The therapeutic landscape for engineered antibodies is experiencing unprecedented growth, driven by advances in protein engineering that enable highly targeted and potent therapeutic modalities. The commercial pipeline for bispecific antibodies (BsAbs) and Antibody-Drug Conjugates (ADCs) is expanding rapidly, reflecting their significant clinical and commercial potential [98] [99].
Table 1: Global Market Forecast for Engineered Antibody Therapeutics
| Therapeutic Modality | Market Size (2024/2025) | Projected Market Size | Projected CAGR | Key Growth Drivers |
|---|---|---|---|---|
| Bispecific Antibodies (Global) | USD 7.49 billion (2024) [98] | USD 76.67 billion by 2032 [98] | 33.72% (2025-2032) [98] | Rising cancer incidence, demand for targeted therapy, rapid regulatory approvals [100] [98] |
| Bispecific Antibodies (U.S.) | USD 11.97 billion (2024) [99] | USD 448.62 billion by 2035 [99] | 44.52% (2026-2035) [99] | New product approvals, expansion into non-oncology diseases, manufacturing innovations [99] |
| Antibody-Drug Conjugates (Global) | - | USD 16+ billion (expected 2025 full-year sales) [101] | - | Precision targeting, potent cytotoxicity, expanding indications and targets [101] [49] |
The pipeline diversity is another key indicator of market vitality. The global ADC landscape is particularly robust, with over 200 candidates in clinical development and 41 ADCs in Phase III trials as of 2025, targeting more than 50 different antigens [101] [49]. The bsAb pipeline is similarly strong, with clinical activity dominated by Phase I/I-II trials (approximately 35% share), though the late-stage segment is expected to grow the fastest, fueled by successful commercialization and accelerated regulatory approvals [99].
Key Therapeutic Platforms and Engineering Trends
Bispecific Antibodies (BsAbs)
BsAbs are synthetic molecules engineered to bind two distinct antigens or epitopes simultaneously. This capability extends beyond the simple additive effect of two monoclonal antibodies, enabling novel mechanisms of action such as the recruitment of immune cells to tumor sites [100].
Table 2: Key Formats and Applications of Bispecific Antibodies
| Format/Architecture | Key Features | Dominant Applications | Examples (Approved) |
|---|---|---|---|
| IgG-like BsAbs (with Fc region) | Longer half-life (FcRn-mediated recycling), higher stability, retained Fc-mediated effector functions (ADCC, CDC, ADCP) [100]. | Oncology, Autoimmune diseases | Mosunetuzumab, Glofitamab [100] [99] |
| Non-IgG-like BsAbs (without Fc region) | Smaller size, improved tissue penetration, avoids Fc-mediated toxicity, shorter half-life [100]. | Hematological malignancies | Blinatumomab (a BiTE) [100] |
| T-cell Engagers (TCEs) e.g., BiTEs, DARTs | Bridge T-cells (via CD3) to tumor cells, inducing cytotoxic synapse independent of MHC [102] [100]. | Multiple Myeloma, Lymphoma | Teclistamab, Epcoritamab [99] |
A major trend in bsAb engineering is the sophisticated modification of the Fc region to fine-tune effector functions, extend serum half-life, and improve safety profiles. Fc engineering is a critical lever for controlling immune activation and pharmacokinetics in next-generation bsAbs [103]. Furthermore, the integration of Artificial Intelligence (AI) is accelerating the discovery and design of complex antibody structures, optimizing lead selection, predicting immunogenicity, and streamlining protein engineering [99].
Antibody-Drug Conjugates (ADCs)
ADCs are a class of biopharmaceuticals designed as "magic bullets," comprising a monoclonal antibody covalently linked to a potent cytotoxic payload via a chemical linker [49]. Their development has progressed through multiple generations, each improving upon the last:
- First-Generation: Utilized murine mAbs and unstable linkers, leading to immunogenicity and off-target toxicity [49].
- Second-Generation: Incorporated humanized mAbs and more stable linkers, improving targeting and safety (e.g., Trastuzumab Emtansine) [49].
- Third-Generation: Feature site-specific conjugation and fully human mAbs for improved homogeneity and reduced immunogenicity (e.g., Enfortumab Vedotin) [49].
- Fourth-Generation: Characterized by high Drug-to-Antibody Ratios (DAR ~8) and novel payloads, significantly enhancing antitumor efficacy (e.g., Trastuzumab Deruxtecan) [49].
Table 3: Evolution of ADC Payloads and Linkers
| Component | Traditional/Previous Standard | Next-Generation Trends |
|---|---|---|
| Payloads | Microtubule disruptors (Auristatins MMAE/MMAF, Maytansinoids DM1/DM4) [104] [101]. | Topoisomerase I inhibitors (DXd-deruxtecan) [104] [101]. DNA alkylating agents (PBD dimers) [49]. |
| Linkers | Cleavable (e.g., pH-sensitive) and non-cleavable linkers [49]. | Enzyme-cleavable peptide linkers with high plasma stability; peptide linkers with β-alanine spacers to reduce hydrophobicity [101]. |
The ADC target landscape is also diversifying. While HER2 remains a primary target, new antigens such as HER3, B7-H3, B7-H4, and CLDN18.2 are prominent in late-stage pipelines. A significant emerging trend is the development of bispecific ADCs, which target two antigens to address tumor heterogeneity and resistance mechanisms [101].
Expansion into Non-Oncology Applications
While oncology remains the dominant indication, accounting for approximately 68% of the bsAb market [99], both BsAbs and ADCs are increasingly being explored for applications in autoimmune, inflammatory, and infectious diseases.
For BsAbs, the ophthalmology/rare diseases and immuno-inflammation/autoimmunity segments are expected to grow at the fastest CAGRs [99]. This expansion is driven by the ability of BsAbs to modulate multiple dysregulated signaling pathways simultaneously, offering a powerful approach for complex diseases like rheumatoid arthritis and inflammatory bowel disease (IBD) [102]. AI platforms are being leveraged to discover new bispecific molecules for these inflammatory conditions [102].
The ADC field is also beginning to explore beyond oncology. Clinical trials are investigating ADCs for non-oncology indications such as graft-versus-host disease (GVHD) [101], and research is ongoing for their potential use in treating persistent bacterial infections [49]. This expansion is facilitated by advancements in antibody engineering and screening technologies that improve the safety and stability of these complex molecules [101].
Experimental Protocols for Evaluation
Protocol 1: In Vitro Potency and Specificity Assay for T-cell Engager Bispecifics
This protocol assesses the ability of a T-cell engaging bsAb to mediate specific lysis of target tumor cells.
1. Key Reagents:
- Effector Cells: Peripheral Blood Mononuclear Cells (PBMCs) from healthy donors, isolated via density gradient centrifugation (e.g., Ficoll-Paque).
- Target Cells: Tumor cell lines expressing the target tumor-associated antigen (TAA) and antigen-negative control cell lines.
- Bispecific Antibody: Serial dilutions of the T-cell engager (e.g., anti-CD3 x anti-TAA).
- Detection Reagent: Lactate Dehydrogenase (LDH) release assay kit or flow cytometry with Annexin V/PI staining for apoptosis.
2. Methodology:
- Co-culture Setup: Seed target cells in a 96-well plate. Add PBMCs at a pre-optimized Effector:Target (E:T) ratio (e.g., 10:1). Treat with serial dilutions of the bsAb. Include controls (target cells alone, target+PBMCs, etc.).
- Incubation: Incubate for 24-48 hours at 37°C, 5% CO₂.
- Cytotoxicity Measurement:
- LDH Assay: Centrifuge plates, collect supernatant, and measure LDH activity per kit instructions. Calculate % specific lysis:
[(Experimental LDH - Spontaneous LDH) / (Maximum LDH - Spontaneous LDH)] * 100. - Flow Cytometry: Harvest co-culture cells, stain with Annexin V and Propidium Iodide (PI), and analyze by flow cytometry to quantify apoptotic and dead target cells.
- LDH Assay: Centrifuge plates, collect supernatant, and measure LDH activity per kit instructions. Calculate % specific lysis:
- Data Analysis: Generate dose-response curves and calculate the half-maximal effective concentration (ECâ‚…â‚€) for bsAb-induced cytotoxicity.
This assay validates the core mechanism of action of TCEs, which is to bridge T-cells and tumor cells, leading to T-cell activation and targeted tumor cell killing, a mechanism highlighted in recent reviews [100].
Protocol 2: ADC Internalization and Bystander Killing Effect Assay
This protocol evaluates the internalization efficiency of an ADC and the subsequent cytotoxic effect on both antigen-positive and antigen-negative neighboring cells.
1. Key Reagents:
- ADC Conjugate: The ADC of interest, preferably conjugated with a fluorescent payload or using a pH-sensitive dye (e.g., pHrodo) labeled on the antibody.
- Cell Lines: Target antigen-positive tumor cells, isogenic target antigen-negative tumor cells, and a co-culture of both.
- Payload Metabolite: The membrane-permeable cytotoxic payload (e.g., DXd for Topoisomerase I inhibitor payloads).
2. Methodology:
- Internalization Assay:
- Seed antigen-positive cells. The next day, add the fluorescently labeled ADC.
- Incubate for various time points (e.g., 0, 2, 4, 8, 24h) at 37°C.
- Analyze cells by flow cytometry or confocal microscopy to track the internalization of the fluorescent signal into acidic compartments (lysosomes).
- Bystander Killing Assay:
- Setup A (Conditioned Media): Treat antigen-positive cells with the ADC for 24h. Collect the conditioned media, filter to remove cells and debris, and apply it to a monolayer of antigen-negative cells.
- Setup B (Direct Co-culture): Co-culture antigen-positive and antigen-negative cells (e.g., at a 1:1 ratio) and treat with the ADC.
- Incubation and Analysis: After 72-96 hours, measure cell viability using a cell viability assay (e.g., CellTiter-Glo). A significant reduction in viability of antigen-negative cells in both setups indicates a potent bystander effect.
- Internalization Assay:
The bystander effect, where the released payload diffuses out of the target cell to kill adjacent cells, is a critical feature of modern ADCs, especially those with topoisomerase I inhibitor payloads like deruxtecan (DXd) [49]. This mechanism is particularly effective against tumors with heterogeneous antigen expression [49].
The Scientist's Toolkit: Key Research Reagent Solutions
Table 4: Essential Research Reagents for Engineered Antibody Development
| Research Reagent / Material | Function in R&D |
|---|---|
| BEAT Protein Platform (IGI Therapeutics) | A proprietary protein platform used for developing investigational bsAbs for oncology and autoimmune diseases [102]. |
| DuoBody Platform (Genmab) | A validated technology platform for the efficient and scalable generation of IgG-like bsAbs [98]. |
| Knobs-into-Holes (KiH) Technology | An Fc engineering strategy that ensures correct heavy chain pairing, minimizing homodimer formation and improving bsAb yield [100] [98]. |
| PEG-based Linkers (e.g., from Biopharma PEG) | Polyethylene glycol (PEG) linkers are used as building blocks in ADC development to improve solubility, stability, and pharmacokinetic properties [101]. |
| Stable Tumor Cell Lines (e.g., CHO-K1, Jurkat with engineered antigen expression) | Essential for in vitro functional assays to test the efficacy, specificity, and cell-bridging capabilities of bsAbs and ADCs under controlled conditions [100]. |
Visualizing Core Mechanisms and Workflows
Bispecific T-cell Engager Mechanism
ADC Mechanism of Action and Bystander Effect
Protein engineering is undergoing a transformative revolution, moving from traditional iterative methods to fully autonomous systems that integrate artificial intelligence, robotic automation, and continuous evolution. This paradigm shift is particularly impactful for therapeutic antibody research, where the complexity of disease targets demands increasingly sophisticated molecular solutions. The convergence of AI-driven design tools and self-driving laboratories has created an unprecedented capacity to explore the protein sequence-function landscape systematically, accelerating the development of next-generation biotherapeutics with enhanced specificity, potency, and developability profiles [105] [37].
Traditional protein engineering approaches, including directed evolution and rational design, have yielded remarkable successes in therapeutic antibody development but face inherent limitations in throughput, scalability, and ability to navigate complex fitness landscapes. The emergence of autonomous protein engineering represents a fundamental change in this paradigm, enabling closed-loop design-build-test-learn cycles that operate with minimal human intervention. These integrated systems leverage machine learning algorithms to predict protein behavior, automated laboratory platforms to execute experiments, and continuous analysis to refine design hypotheses, thereby compressing development timelines from years to months while accessing previously unexplored regions of the protein functional universe [37] [106].
Application Notes: Core Platforms and Technologies
Autonomous Laboratory Platforms for Continuous Evolution
Recent advancements in laboratory automation have enabled the creation of integrated systems capable of continuous protein evolution. The iAutoEvoLab platform represents a state-of-the-art example, featuring high-throughput capabilities, enhanced reliability, and the ability to operate autonomously for extended periods (approximately one month). This system addresses critical bottlenecks in conventional protein engineering by combining programmable genetic circuits with growth-coupled selection systems that link protein function to cellular survival [37].
Table 1: Key Characteristics of Autonomous Protein Evolution Platforms
| Platform Feature | Technical Specification | Therapeutic Antibody Application |
|---|---|---|
| Operational Duration | ~1 month autonomous operation | Enables continuous maturation of antibody affinity |
| Genetic System | OrthoRep with engineered genetic circuits | Allows continuous evolution of full-length IgGs |
| Selection Strategy | Growth-coupled dual selection | Simultaneous optimization of affinity and specificity |
| Throughput Capacity | 100-1000x conventional directed evolution | Comprehensive exploration of CDR mutant libraries |
| Integration Level | Fully automated design-build-test-learn cycle | Closed-loop optimization of developability parameters |
These platforms employ sophisticated genetic circuits for selective pressure, such as the NIMPLY circuit for operator selectivity and dual-selection systems for sensitivity tuning. For antibody engineering, this enables simultaneous optimization of multiple parameters, including target binding affinity, specificity against off-target antigens, and stability under physiological conditions. The platform has demonstrated capability to evolve proteins from inactive precursors to fully functional entities, as evidenced by the development of CapT7, a T7 RNA polymerase fusion protein with mRNA capping activity that can be directly applied to in vitro transcription systems for mRNA therapeutic production [37].
AI-Driven Protein Design Framework
The integration of artificial intelligence has transformed protein engineering from an empirical art to a systematic engineering discipline. A comprehensive seven-toolkit workflow has emerged as the foundational framework for AI-driven protein design, organizing disparate computational tools into a coherent pipeline from concept to validation [106]:
- Protein Database Search (T1): Identification of structural homologs and sequence templates for initial scaffold design
- Protein Structure Prediction (T2): High-accuracy folding prediction using AlphaFold2 and related tools
- Protein Function Prediction (T3): Annotation of potential function and binding sites
- Protein Sequence Generation (T4): Creation of novel sequences matching desired structural or functional constraints
- Protein Structure Generation (T5): De novo backbone design for entirely new folds and topologies
- Virtual Screening (T6): Computational assessment of stability, affinity, and immunogenicity
- DNA Synthesis & Cloning (T7): Translation of optimized designs into DNA sequences for expression
This framework has been successfully validated through multiple applications, including the AI-guided evolution of β-lactamase where mutation suggestions from tools T3 and T6 accelerated discovery of drug-resistant variants, and the de novo creation of a COVID-19 binding protein through combination of structure generation (T5), sequence design (T4), and virtual screening (T6) [106].
Next-Generation Therapeutic Modalities
Autonomous protein engineering is enabling the development of novel therapeutic modalities beyond conventional monoclonal antibodies. These next-generation modalities address longstanding challenges in oncology, autoimmune disorders, and infectious diseases through innovative molecular designs:
Bispecific T-Cell Engagers (BiTEs): These engineered antibodies simultaneously target tumor-associated antigens and CD3 receptors on T-cells, effectively redirecting immune effector cells to malignant cells. Blinatumomab represents a clinically validated example that has demonstrated remarkable efficacy in B-cell malignancies [107].
Antibody-Drug Conjugates (ADCs): Autonomous platforms optimize the antibody component for enhanced tumor specificity and conjugation compatibility, while also engineering the linker chemistry for controlled payload release. Next-generation ADCs benefit from AI-driven prediction of immunogenicity and stability parameters [107] [108].
Cell Therapies with Engineered Receptors: Chimeric antigen receptor T-cell (CAR-T) therapies and T-cell receptor (TCR) therapies incorporate engineered targeting domains optimized for specific antigen recognition while minimizing off-target effects. Automated platforms enable rapid iteration of extracellular domain designs [108].
PROTACs and Molecular Degraders: Though not antibody-based, these modalities leverage protein engineering principles to create bifunctional molecules that recruit cellular machinery for targeted protein degradation. Autonomous engineering accelerates the optimization of binding domains for both the target protein and degradation machinery [108].
Table 2: Next-Generation Therapeutic Modalities Enabled by Autonomous Protein Engineering
| Modality Class | Key Engineering Parameters | Therapeutic Application |
|---|---|---|
| Bispecific Antibodies | Cross-arm specificity, geometric orientation, Fc engineering | Oncology, immune disorders |
| Antibody-Drug Conjugates | Linker stability, drug-antibody ratio, internalization efficiency | Solid tumors, hematologic malignancies |
| CAR-T/TCR Therapies | Binding affinity, signaling domains, safety switches | Hematologic cancers, solid tumors |
| Fusion Proteins | Half-life extension, receptor affinity, valency control | Cytokine modulation, receptor blockade |
| De Novo Enzymes | Catalytic efficiency, substrate specificity, cofactor requirement | Metabolic disorders, prodrug activation |
Experimental Protocols
Protocol: Automated Continuous Evolution for Antibody Affinity Maturation
This protocol describes the implementation of an autonomous continuous evolution system for antibody affinity maturation, adapted from the iAutoEvoLab platform with specific modifications for therapeutic antibody engineering [37].
Materials and Reagents
- OrthoRep plasmid system (or similar orthogonal replication system)
- Growth-coupled selection circuit (e.g., tetA/GFP dual reporter system)
- Yeast surface display platform (e.g., Aga2p fusion system)
- Automated bioreactor array with continuous culture capability
- FACS system with integrated sampling
- Antigen conjugates for selection pressure (biotinylated for magnetic separation)
- NGS library preparation reagents
Procedure
Library Design and Construction
- Amplify antibody variable regions with error-prone PCR (targeting 0.5-1 mutation/kb)
- Clone into yeast surface display vector with C-terminal epitope tag
- Electroporate library into yeast strain harboring OrthoRep system
- Determine library diversity by plating and colony counting (target >10^9 transformants)
System Configuration and Calibration
- Program genetic circuit parameters for selective pressure
- Establish baseline growth rates in absence of selection
- Titrate selection pressure to achieve 50-70% growth inhibition
- Validate display efficiency by FACS analysis (>90% display required)
Continuous Evolution Operation
- Initiate continuous culture in automated bioreactors
- Maintain culture at mid-log phase (OD600 = 0.5-0.8) via automated dilution
- Apply periodic antigen pressure cycles (24-48 hour intervals)
- Monitor population dynamics via daily sampling and FACS analysis
- Implement "selection holidays" to prevent evolutionary dead ends
Variant Isolation and Characterization
- After 4-6 weeks of continuous evolution, harvest population
- Isolate individual clones by FACS sorting based on binding signal
- Sequence variants using the every variant sequencing (evSeq) pipeline
- Characterize affinity improvements via SPR or BLI
Critical Parameters
- Maintain effective population size >10^7 to prevent bottleneck effects
- Monitor mutation rate to ensure optimal diversity (target 10^-5 - 10^-4 mutations/base/generation)
- Adjust selection stringency progressively as affinity improves
- Include neutral diversity controls to assess baseline mutation rate
Protocol: AI-Guided De Novo Antibody Design
This protocol outlines the implementation of the seven-toolkit AI framework for de novo antibody design, specifically for creating antibodies targeting complex epitopes [106].
Materials and Reagents
- Structure prediction software (AlphaFold2, ESMFold)
- Sequence design tools (ProteinMPNN, Rosetta)
- Structure generation platforms (RFDiffusion, Chroma)
- Virtual screening pipelines (docking software, molecular dynamics)
- Mammalian expression system (HEK293 or CHO cells)
- ELISA kits for binding validation
- SPR or BLI instrument for affinity measurement
Procedure
Target Characterization (T1-T3)
- Identify known structural homologs using PDB search (T1)
- Predict target structure and conformational states (T2)
- Map functional epitopes and conserved residues (T3)
- Define design constraints based on target biology
Scaffold Generation and Selection (T4-T5)
- Generate novel backbone architectures using RFDiffusion (T5)
- Design complementary binding surfaces for target epitope
- Generate sequence variants using ProteinMPNN (T4)
- Filter designs by structural integrity and novelty
In Silico Validation (T6)
- Perform molecular docking with target antigen
- Run short MD simulations to assess stability
- Predict developability parameters (solubility, viscosity)
- Rank candidates by composite fitness score
Experimental Validation (T7)
- Synthesize top 50-100 designs as IgG constructs (T7)
- Express in mammalian system and purify via protein A
- Screen for binding via ELISA against target antigen
- Characterize affinity of positive hits via SPR/BLI
- Assess specificity against antigen family members
Critical Parameters
- Balance novelty with foldability in scaffold design
- Maintain CDR structural compatibility with human germline
- Include humanization checkpoints throughout process
- Prioritize designs with favorable developability profiles
Workflow Visualization
AI-Driven Autonomous Protein Engineering Workflow
The Scientist's Toolkit: Essential Research Reagents and Platforms
Table 3: Key Research Reagent Solutions for Autonomous Protein Engineering
| Research Tool | Function/Application | Implementation in Therapeutic Antibody Engineering |
|---|---|---|
| OrthoRep System | Orthogonal DNA replication system for continuous in vivo evolution | Enables continuous mutagenesis of full-length antibody genes in yeast |
| ProteinMPNN | Neural network for protein sequence design | Designs humanized antibody sequences for given backbone structures |
| RFDiffusion | Generative model for novel protein backbone creation | Creates novel antibody scaffolds for targeting complex epitopes |
| Yeast Surface Display | Platform for antibody expression and screening | Enables FACS-based selection of high-affinity antibody variants |
| evSeq/LevSeq | Every variant sequencing technology | Provides complete sequence-function data for entire mutant libraries |
| Autoinduction Systems | Automated induction of protein expression in high-throughput | Facilitates parallel expression of hundreds of antibody variants |
| Cell-Free Expression | Transcription/translation without living cells | Rapid screening of antibody variants directly from DNA designs |
| SPR/BLI Platforms | Label-free binding affinity and kinetics measurement | Characterizes antibody-antigen interactions with high precision |
Conclusion
The field of protein engineering for therapeutic antibodies is undergoing a profound transformation, driven by the convergence of computational biology, AI, and high-throughput experimental methods. The integration of tools like RFDiffusion and ProteinMPNN with traditional methodologies has created a powerful, iterative design cycle that accelerates the development of novel biologics. Key takeaways include the critical need to address developability challenges early and the expanding potential of engineered formats like bispecifics and ADCs. Future progress will hinge on leveraging autonomous discovery platforms and AI to navigate the complex sequence-function landscape, ultimately enabling the precise design of next-generation antibody therapeutics for increasingly complex diseases. This synergy between computation and experiment promises to unlock new therapeutic modalities and redefine the standard of care in clinical medicine.
References
- 1. Biologics Market Size, Share and Trends 2025 to 2034 [https://www.precedenceresearch.com/biologics-market]
- 2. Biologics Manufacturing Market Size to Hit USD 162.51 ... [https://www.biospace.com/press-releases/biologics-manufacturing-market-size-to-hit-usd-162-51-billion-by-2034-growing-at-an-extraordinary-17-1-cagr]
- 3. Biologics Market - Size, Growth, Share & Report [https://www.mordorintelligence.com/industry-reports/biologics-market]
- 4. Biologic Manufacturing: Process, Steps, and Challenges [https://www.susupport.com/blogs/knowledge/biologics-manufacturing-process-steps-challenges]
- 5. Biomanufacturing: How Biologics are Made [https://biotechprimer.com/how-biologics-are-made/]
- 6. Protein engineering approaches for antibody fragments [https://pmc.ncbi.nlm.nih.gov/articles/PMC6426644/]
- 7. Engineering protein-based therapeutics through structural ... [https://www.nature.com/articles/s41467-023-38039-x]
- 8. Technological advancements in antibody-based therapeutics ... [https://jbiomedsci.biomedcentral.com/articles/10.1186/s12929-025-01190-2]
- 9. Protein engineering as a driver of innovation in ... [https://link.springer.com/article/10.1007/s44371-025-00313-w]
- 10. Applying computational protein design to therapeutic ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2025.1571371/full]
- 11. October 2025: Designing Antibodies with Artificial ... [https://www.biointron.com/antibody-industry-trends/october-2025-designing-antibodies-with-artificial-lntelligence.html]
- 12. Market Size, Growth Trends 2026-2035 Antibody Discovery [https://www.researchnester.com/reports/antibody-discovery-market/7087]
- 13. Antibody Discovery Market Size, Share & 2030 Growth ... [https://www.mordorintelligence.com/industry-reports/antibody-discovery-market]
- 14. Simplifying complex antibody engineering using machine ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10733906/]
- 15. Machine learning optimization of candidate antibody yields ... [https://www.nature.com/articles/s41467-023-39022-2]
- 16. Highly accurate protein structure prediction with AlphaFold [https://www.nature.com/articles/s41586-021-03819-2]
- 17. Computational approaches to therapeutic antibody design [https://pmc.ncbi.nlm.nih.gov/articles/PMC7947987/]
- 18. Study sets new data benchmark for trustworthy antibody AI [https://www.stats.ox.ac.uk/news/study-sets-new-data-benchmark-trustworthy-antibody-ai]
- 19. Study Finds Generative AI Outperforms Nature in Protein ... [https://www.biopharminternational.com/view/study-finds-generative-ai-outperforms-nature-in-protein-design]
- 20. De novo design of protein structure and function with ... [https://www.nature.com/articles/s41586-023-06415-8]
- 21. Atomic context-conditioned protein sequence design using ... [https://www.nature.com/articles/s41592-025-02626-1]
- 22. Atomically accurate de novo design of antibodies with ... [https://www.nature.com/articles/s41586-025-09721-5]
- 23. A novel decoding strategy for ProteinMPNN to design with ... [https://www.sciencedirect.com/science/article/pii/S2001037025003198]
- 24. Scientists use AI to create antibodies entirely from scratch [https://www.drugtargetreview.com/news/190363/scientists-use-ai-to-create-antibodies-entirely-from-scratch/]
- 25. Rational Protein Design [https://www.biosynsis.com/rational-protein-design.html]
- 26. Insights Into Protein Engineering: Methods and Applications [https://www.the-scientist.com/insights-into-protein-engineering-methods-and-applications-72286]
- 27. 4D structural biology–quantitative dynamics in the ... [https://www.nature.com/articles/s41467-025-62982-6]
- 28. Quantitative analysis of protein dynamics using a deep ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10941960/]
- 29. Site-directed mutagenesis method for protein engineering ... [https://www.sciencedirect.com/science/article/abs/pii/S0006291X21004034]
- 30. A Technical Guide to Directed Evolution for Enhancing ... [https://www.ranomics.com/a-technical-guide-to-directed-evolution-for-enhancing-protein-stability-and-function]
- 31. A primer to directed evolution: current methodologies and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10074555/]
- 32. Antibodies 101: Antibody Engineering and Directed Evolution [https://blog.addgene.org/antibodies-101-antibody-engineering-and-directed-evolution]
- 33. Combination of error-prone PCR (epPCR) and Circular ... [https://www.nature.com/articles/s41598-024-66584-y]
- 34. High-throughput strategies for monoclonal antibody screening [https://jbioleng.biomedcentral.com/articles/10.1186/s13036-025-00513-z]
- 35. Pioneer: a synthetic human antibody phage display library ... [https://pubmed.ncbi.nlm.nih.gov/40808547/]
- 36. Growth-coupled continuous directed evolution by MutaT7 ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12016552/]
- 37. An industrial automated laboratory for programmable ... [https://www.nature.com/articles/s44286-025-00303-w]
- 38. Advancements in mammalian display technology for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11459229/]
- 39. Advancements in mammalian display technology for ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2024.1469329/full]
- 40. In vitro display technologies in antibody discovery and engineering... [https://pipebio.com/blog/antibody-display-technologies]
- 41. Development of a novel mammalian display system ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7939478/]
- 42. Bispecific Antibodies: From Research to Clinical Application [https://pmc.ncbi.nlm.nih.gov/articles/PMC8131538/]
- 43. Bispecific antibodies: unleashing a new era in oncology ... [https://molecular-cancer.biomedcentral.com/articles/10.1186/s12943-025-02390-y]
- 44. Antibody drug conjugate: the “biological missile†... - Nature [https://www.nature.com/articles/s41392-022-00947-7]
- 45. Antibody–drug conjugates in cancer therapy: mechanisms ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11283588/]
- 46. Roles of Fc-Mediated Effector Functions in Broadly ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7748341/]
- 47. Fc-dependent antibody effector functions in SARS-CoV-2 ... [https://www.nature.com/articles/s41577-022-00813-1]
- 48. Fc-Mediated Antibody Effector Functions During ... - Frontiers [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2019.00548/full]
- 49. Antibody–Drug Conjugates (ADCs): current and future ... [https://jhoonline.biomedcentral.com/articles/10.1186/s13045-025-01704-3]
- 50. Antibody–drug conjugates: Recent advances in payloads [https://www.sciencedirect.com/science/article/pii/S2211383523002320]
- 51. Fc-Engineered Antibodies with Enhanced Fc-Effector ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7603375/]
- 52. Boosting therapeutic potency of antibodies by taming Fc ... [https://www.nature.com/articles/s12276-019-0345-9]
- 53. Modifying antibody-FcRn interactions to increase the transport ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10312019/]
- 54. Dual Fc optimization to increase the cytotoxic activity of a ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2022.957874/full]
- 55. Antibody Fc engineering improves frequency and promotes ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4239333/]
- 56. Asymmetrical Fc Engineering Greatly Enhances Antibody ... [https://www.sciencedirect.com/science/article/pii/S0021925819747474]
- 57. Prediction of human pharmacokinetics of Fc-engineered ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11938312/]
- 58. Engineering FcRn binding kinetics dramatically extends ... [https://jbioleng.biomedcentral.com/articles/10.1186/s13036-025-00506-y]
- 59. Fc-engineered large molecules targeting blood-brain ... [https://www.nature.com/articles/s41467-025-57108-x]
- 60. Antibody Humanization Strategies, Challenges, and ... [https://www.rapidnovor.com/antibody-humanization-strategies-challenges-innovations/]
- 61. Deimmunization of protein therapeutics – Recent advances ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7779837/]
- 62. Machine-guided dual-objective protein engineering for ... [https://www.sciencedirect.com/science/article/abs/pii/S2405471225001322]
- 63. The immunogenicity of humanized and fully human antibodies [https://pmc.ncbi.nlm.nih.gov/articles/PMC2881252/]
- 64. Humanization of antibodies using a machine learning ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8760955/]
- 65. Comparing Immune-Humanized Mouse Platforms for ... [https://resources.jax.org/immuno-oncology-oncology/comparing-immune-humanized-mouse-platforms-for-preclinical-applications-10222024]
- 66. The Immune Signatures data resource, a compendium of ... [https://www.nature.com/articles/s41597-022-01714-7]
- 67. Automated optimisation of solubility and conformational ... [https://www.nature.com/articles/s41467-023-37668-6]
- 68. The aggregation challenge: Measuring stability in ... [https://www.ddw-online.com/the-aggregation-challenge-measuring-stability-in-therapeutic-antibodies-34398-202504/]
- 69. Enhancing the Thermostability and solubility of a single- ... [https://pubmed.ncbi.nlm.nih.gov/39961023/]
- 70. Enhancing therapeutic antibody production through amino ... [https://www.frontiersin.org/journals/bioengineering-and-biotechnology/articles/10.3389/fbioe.2025.1567923/full]
- 71. Impact of Monoclonal Antibody Aggregates on Effector ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12015860/]
- 72. Impact of antibody Fc engineering on translational ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12239809/]
- 73. May 2025: Fc Engineering - Innovations in Antibody ... [https://www.biointron.com/antibody-industry-trends/may-2025-fc-engineering-innovations-in-antibody-therapeutics.html]
- 74. Week 1, April 2025: Emerging Trends in Fc Domain ... [https://www.biointron.com/antibody-industry-trends/week-1-april-2025-emerging-trends-in-fc-domain-engineering-for-therapeutic-development.html]
- 75. Why Biopharma Companies are Prioritizing Affinity ... [https://precisionantibody.com/why-biopharma-companies-are-prioritizing-affinity-maturation-2025/]
- 76. Antibody Affinity Maturation: Review, Strategies, and Process [https://www.sinobiological.com/resource/antibody-technical/antibody-affinity-maturation]
- 77. The Critical Role of Residues on CDR Loops of Antibodies ... [https://www.mdpi.com/1420-3049/30/3/532]
- 78. Understanding the Significance and Implications of ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2018.02278/full]
- 79. 50 Years of Antibody Numbering Schemes: A Statistical ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11672675/]
- 80. Antibody numbering schemes: advances, comparisons and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11997657/]
- 81. How Your 2025 Agenda Addresses the Big Industry ... [https://informaconnect.com/antibody-engineering-therapeutics/how-your-2025-agenda-addresses-the-big-industry-challenges/]
- 82. A machine learning strategy for the identification of key in ... [https://pubmed.ncbi.nlm.nih.gov/PMC10448975]
- 83. Frontier of therapeutic antibody discovery: The challenges ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3531614/]
- 84. High-throughput strategies for monoclonal antibody screening [https://pmc.ncbi.nlm.nih.gov/articles/PMC12063422/]
- 85. High-Throughput Screening in Protein Engineering [https://pmc.ncbi.nlm.nih.gov/articles/PMC4632782/]
- 86. An Antibody Engineering Platform for High Throughput ... [https://www.genedata.com/resources/learn/details/poster/an-antibody-engineering-platform-for-high-throughput-candidate-screening-engineering-optimization]
- 87. Services for Antibody Engineering [https://www.thermofisher.com/us/en/home/life-science/cloning/gene-synthesis/antibody-engineering-services.html]
- 88. Upcoming Webinars in 2025 [https://www.antibodysociety.org/learningcenter/upcoming-webinars/]
- 89. Antibodies to watch in 2025 [https://pubmed.ncbi.nlm.nih.gov/39711140/]
- 90. Leading Therapeutic Antibodies Approved by FDA 2025 [https://www.genextgenomics.com/top-10-fda-approved-therapeutic-antibodies-you-should-know-in-2025/]
- 91. A comparative study of the developability of full-length ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11457596/]
- 92. In Silico Methods in Antibody Design - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6640671/]
- 93. Antibody Discovery Market Size & Growth Forecast to 2030 [https://www.marketsandmarkets.com/Market-Reports/antibody-discovery-market-182887795.html]
- 94. Evolution of phage display libraries for therapeutic antibody ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10210849/]
- 95. Engineering high affinity antigen-binders [https://www.sciencedirect.com/science/article/abs/pii/S1876162323001268]
- 96. Prediction of human pharmacokinetics of Fc-engineered ... [https://pubmed.ncbi.nlm.nih.gov/40133232/]
- 97. Factors influencing monoclonal antibody pharmacokinetics ... [https://www.sciencedirect.com/science/article/abs/pii/S016836592500803X]
- 98. Bispecific Antibodies Market - Global Forecast 2025-2032, ... [https://finance.yahoo.com/news/bispecific-antibodies-market-global-forecast-082300615.html]
- 99. U.S. Bispecific Antibody Market to Grow at 44.52% till 2035 [https://www.towardshealthcare.com/insights/us-bispecific-antibody-market-sizing]
- 100. Bispecific antibodies: unleashing a new era in oncology ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12320366/]
- 101. ADC Market 2025: Rising Sales and Growing Phase III ... [https://www.biochempeg.com/article/447.html]
- 102. Next-Generation Bispecific Antibody Market Trends and [https://www.globenewswire.com/news-release/2025/09/30/3158764/0/en/Next-Generation-Bispecific-Antibody-Market-Trends-and-Competitive-Landscape-Report.html]
- 103. Antibody Engineering and Therapeutics 2025 [https://www.seromyx.com/antibody-engineering-and-therapeutics-2025]
- 104. The evolving landscape of antibody-drug conjugates ... [https://www.nature.com/articles/s41698-025-01131-0]
- 105. The Role of AI-Driven De Novo Protein Design in the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12467925/]
- 106. Engineering Life: A New Roadmap for AI-Driven Protein ... [https://www.ailurus.bio/post/engineering-life-a-new-roadmap-for-ai-driven-protein-design]
- 107. Protein Engineering Paving the Way for Next-Generation ... [https://www.mdpi.com/2673-8937/5/3/28]
- 108. Next generation therapeutics [https://www.astrazeneca.com/r-d/next-generation-therapeutics.html]